Help Page | ||
| CancerIN is a webserver for the classification of active and inactive anti-cancer compounds. It also allow the user to designing their analogs against cancer cell lines. Three major modules have been integrated to facilitate users. This page is designed for providing help on CancerIN. Please click on topic for detail help. |
| 1. Draw Structure | 2. Virtual Screening | 3.Analog Designing |
| Important Fingerprints |
| We have identified 126 fingerprints/substructures that play an important role in classification of active vs. inactive anti-cancer inhibitors. The fingerprints will help in better understanding of anti-cancer inhibitors. We encourage users to understand these 126 fingerprints, which will help them in all three modules: Draw structure, Virtual Screening and Analog Designing. Click here to view 126 fingerprints in a tabular format |
| 1. Draw Structure |
Here user can draw their query molecules using different options provided in the Marvin applet. This applet is Java based, so Java should be enabled in your browser. After submission, it will return the five best anticancer molecules, which is nearest to the query molecule. User can provide the email address to obtain result on their email id. |
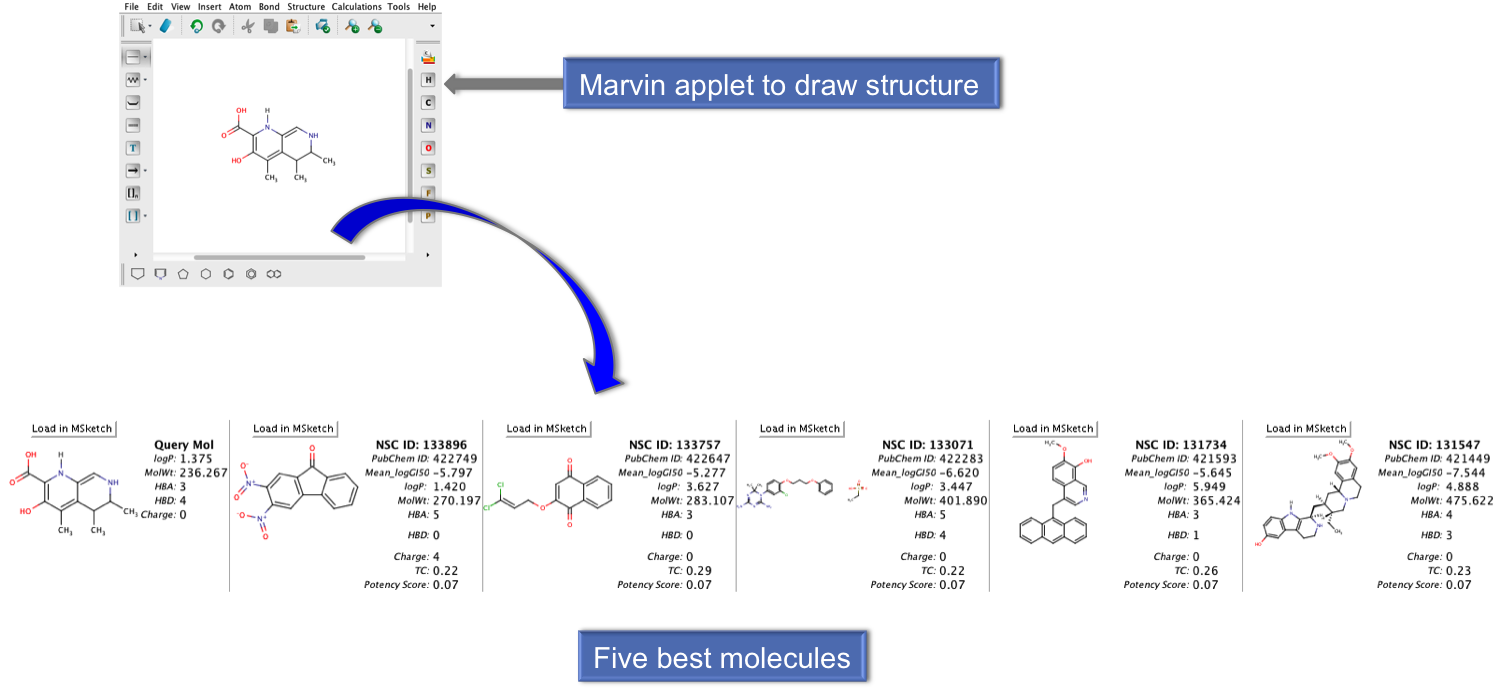 |
| Go to Top |
| Virtual Screening |
In virtual screening user can provide the list of molecules in smile format or they can also upload the file containing molecules. After submission, user will get the best five anticancer molecules for each query molecule. We suggest user to provide thier email address to obtain the results in their email id. |
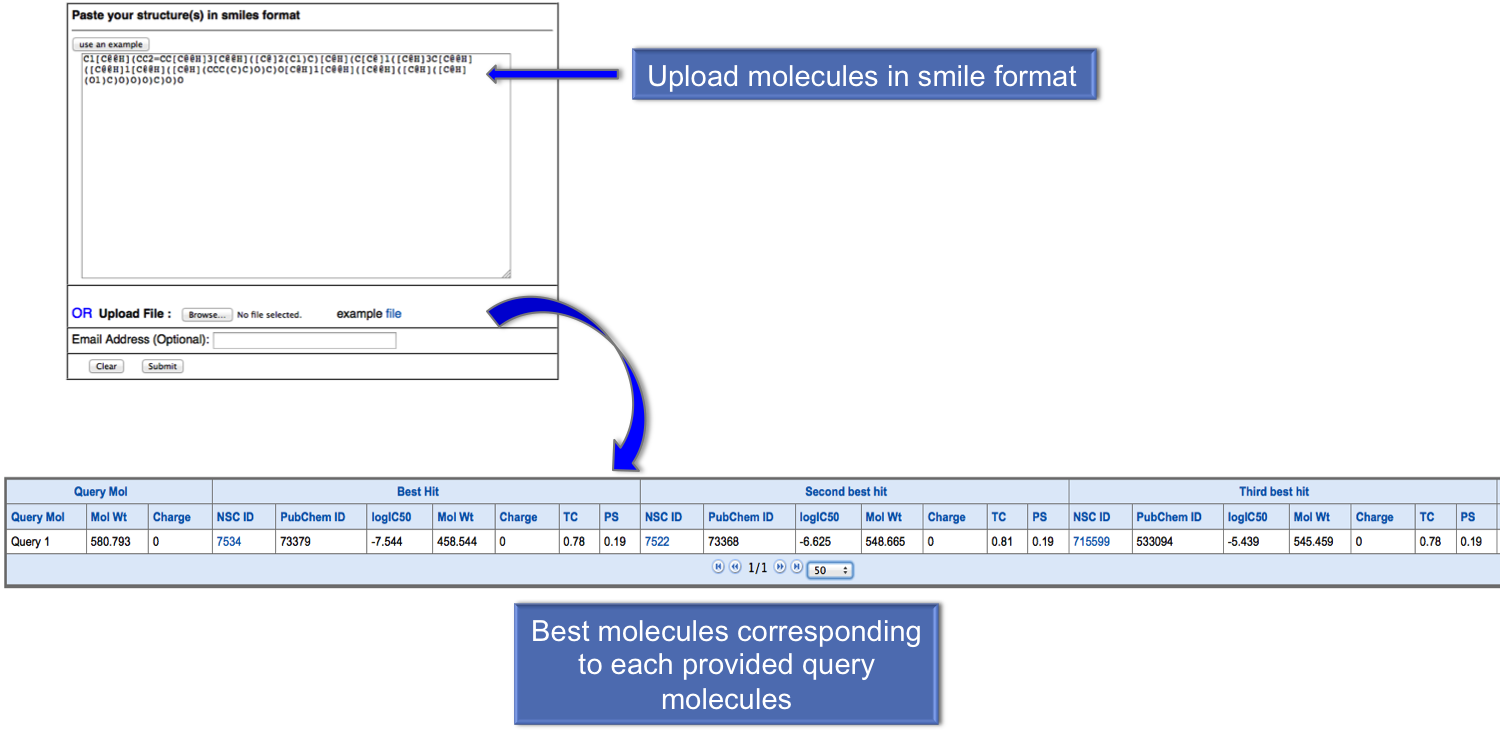 |
| Go to Top |
| Analog Designing |
In this module, first user can build the analog molecules by providing scaffold, building blocks and linkers. After submission, user will get the anticancer molecules nearest to the each analog on the basis of their tanimoto coefficient. |
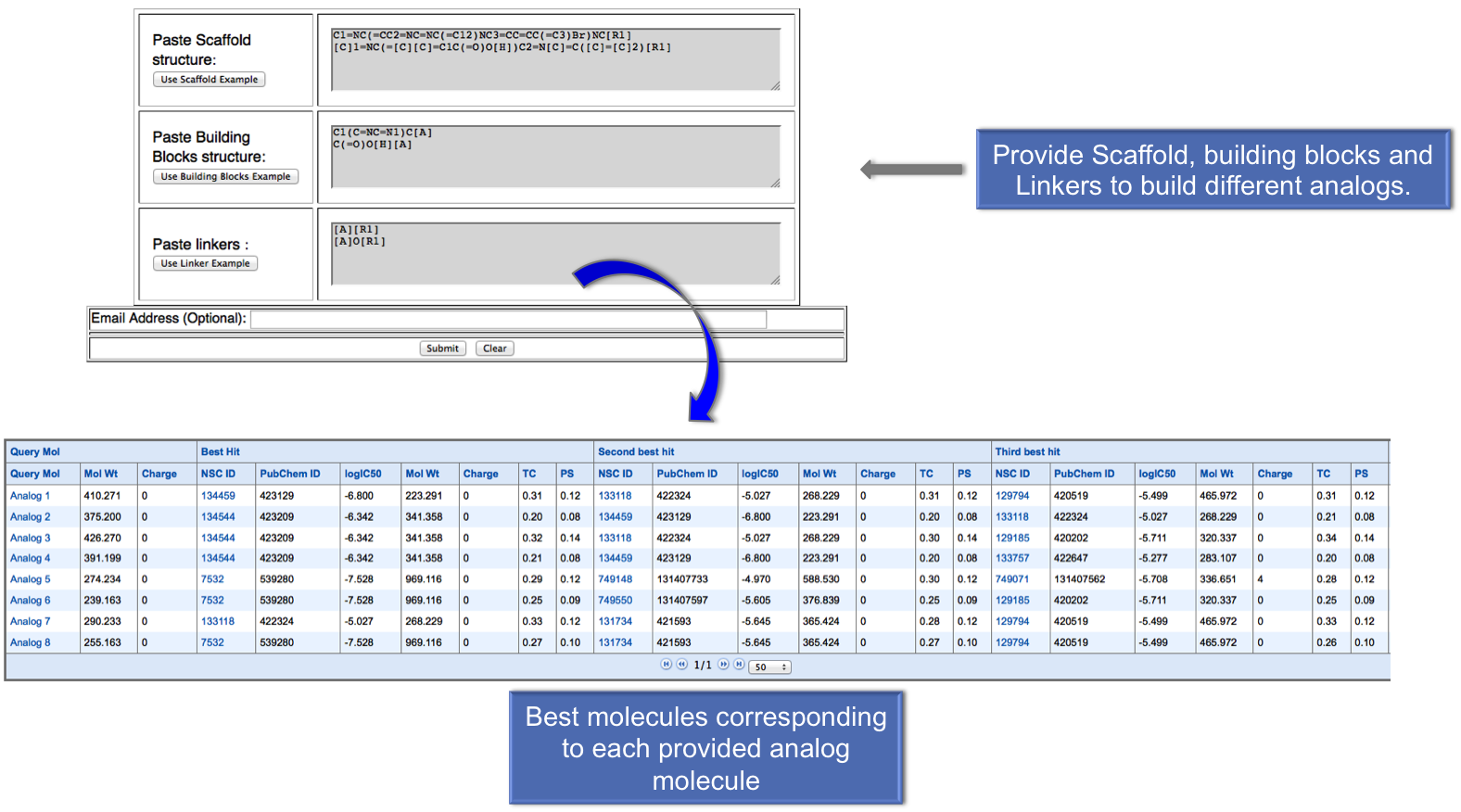 |
| Go to Top |
