Protein Sequence as input to TSP-PRED |
User can enter protein sequences in plain text format (single line) in the input box. User needs to select the method for predicting tertiary structure. (Recommended method: "Both Homology and Abinitio method"). User also needs to input MODELLER key which can be obtained from the MODELLER website (http://salilab.org/modeller/registration.html). Finally a user needs to enter his/her e-mail address where the results of TSP-PRED will be send.
Figure 1 shows how to input the protein sequence in TSP-PRED web server.
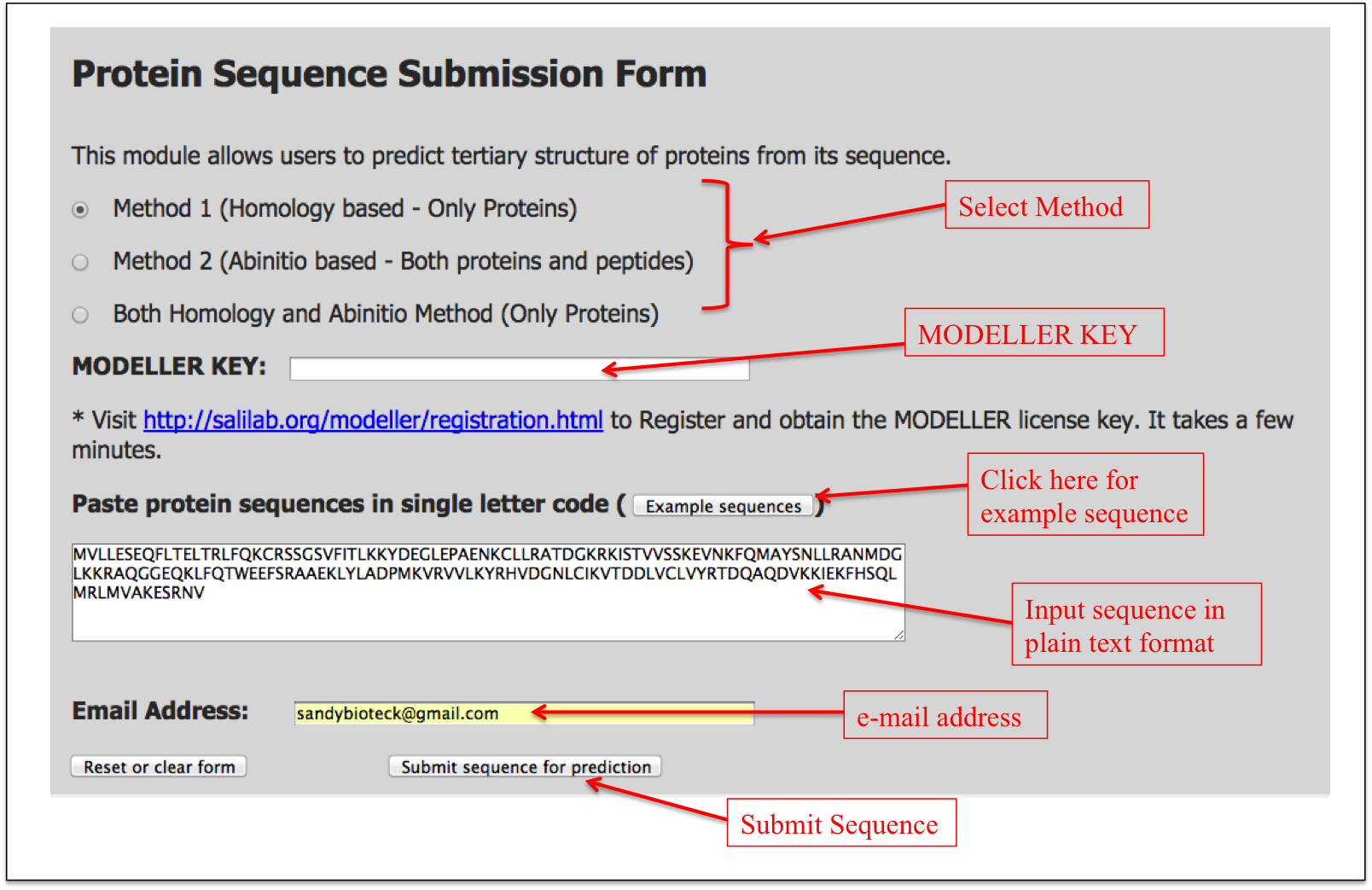
Figure 1. Graphical representation showing how to input the protein sequence in TSP-PRED web server.
Representation of Results |
The results are represented in graphical representation displaying the predicted tertiary structure in Jmol viewer. The structure can also be downloaded in the standard PDB file format. Quality measures of the predicted tertiary structure like Ramachandran Map and Score are also provided in the results. Figure 2 shows the graphical representation of the results obtained using TSP-PRED web server.
