Prediction Performance
Performance Measures
The method is based on the performance of two non-homologous data set of proteins with sequence homology less then 25%.One data set is comprised of 215 proteins (Data sets of PSSM and PSIPRED predicted secondary states) which was also used earlier by Manesh et al (2002) for state prediction of surface accessbility(SA).The other data set is comprised of 502 proteins (Data sets of PSSM and PSIPRED predicted secondary states) obtained from a set of 513 proteins Cuff and Barton (2000) by removing those sequnces which have less then 29 residues.These two data sets have also been used by Ahmad et al (2003)for real value prediction of surface accessibility.
The performance has been assessed for real value prediction of surface accessibility as well as two state (exposed or buried) predictions. For real value prediction of surface accessibility, two parameters have been used
The performance has been assessed for real value prediction of surface accessibility as well as two state (exposed or buried) predictions. For real value prediction of surface accessibility, two parameters have been used
Mean absolute error(MAE)
is defined as the absolute difference between the predicted and experimental(desired) value of relative SA, per residue
| MAE = | 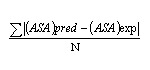 |
Pearson's Correlation Coefficient (r)
is defined as the ratio of the covariance between the predicted and experimental SA values to the product of the standard deviation in the two.
| r = | 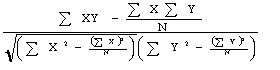 |
where, X is experimental value of relative SA and Y is predicted value of relative SA.
Performace measures for two state predictions
For assessing the quality of two state predictions, three threshold dependent measures have been used.These three parameters can be derived from the four scalar quantities : p (the number of correctly predicted exposed state value); n (the number of correctly predicted burried state values); o (the number of burried state values in correctly predicted as exposed state value); u (the number of exposed state values, incorresctly classified as burried state value).The three paramters derived from these quantities are
| Accuracy = | 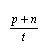 |
| Sn = | 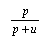 |
| Sp = | 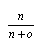 |
Performance
Table 1 The following table shows the Performance of SARpred by using single sequence with and without secondary structure information on both data sets
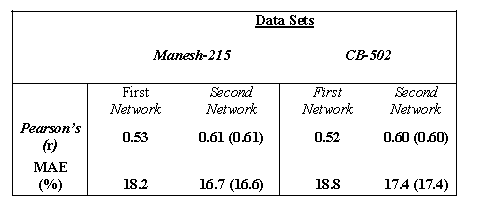
Value in parentheses correspond to the prediction results obtained by excluding the proteins that were used to develop PSIPRED.
Table 2 The following table shows the Performance of SARpred by using multiple sequence with and without secondary structure information on both data sets
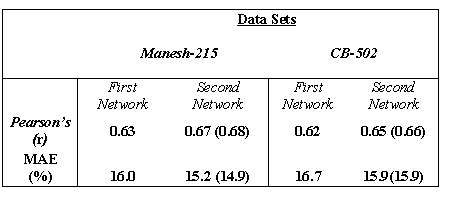
Value in parentheses correspond to the prediction results obtained by excluding the proteins that were used to develop PSIPRED.
Table 3 The following table shows the Performance of SARpred for two states (exposed or buried) prediction by using multiple sequence with and without secondary structure information on Manesh-215 data set
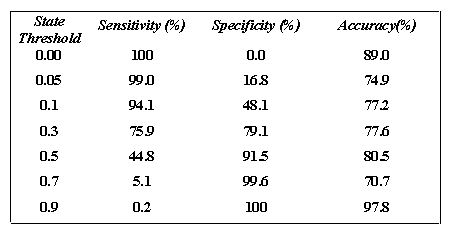
Figure 1 The following figure shows the MAE in various ASA ranges for Manesh-215 data set
The correlation and MAE values obtained in the present study are best among all the previously published real value prediction methods.