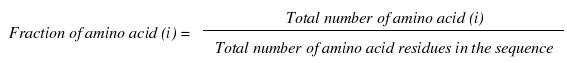ProPrInt: Help page
Following are the detailed results of Support Vector Machine optimization
The result will help you to select optimum "threshold" for the prediction
[A] Escherichia coli
(i)Amino Acid Composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| 1.00 | 0.565 | 1.000 | 0.968 | 1.000 | 0.967 | 0.739 |
| 0.800 | 0.709 | 1.000 | 0.979 | 1.000 | 0.978 | 0.833 |
| 0.600 | 0.811 | 1.000 | 0.986 | 1.000 | 0.985 | 0.894 |
| 0.400 | 0.887 | 1.000 | 0.992 | 1.000 | 0.991 | 0.938 |
| 0.200 | 0.930 | 1.000 | 0.995 | 1.000 | 0.995 | 0.962 |
| 0.000 | 0.957 | 1.000 | 0.997 | 0.999 | 0.997 | 0.976 |
| -0.200 | 0.979 | 1.000 | 0.998 | 0.999 | 0.998 | 0.988 |
| -0.400 | 0.990 | 1.000 | 0.999 | 0.997 | 0.999 | 0.993 |
| -0.600 | 0.994 | 0.999 | 0.999 | 0.994 | 1.000 | 0.994 |
| -0.800 | 0.996 | 0.993 | 0.994 | 0.921 | 1.000 | 0.955 |
| -1.000 | 1.000 | 0.803 | 0.817 | 0.284 | 1.000 | 0.477 |
(ii)Dipeptide Composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| 1.000 | 0.790 | 1.000 | 0.985 | 1.000 | 0.984 | 0.882 |
| 0.800 | 0.884 | 1.000 | 0.992 | 1.000 | 0.991 | 0.936 |
| 0.600 | 0.932 | 1.000 | 0.995 | 1.000 | 0.995 | 0.963 |
| 0.400 | 0.962 | 1.000 | 0.997 | 1.000 | 0.997 | 0.979 |
| 0.200 | 0.981 | 1.000 | 0.999 | 1.000 | 0.998 | 0.989 |
| 0.000 | 0.992 | 1.000 | 0.999 | 1.000 | 0.999 | 0.996 |
| -0.200 | 0.994 | 1.000 | 1.000 | 1.000 | 1.000 | 0.997 |
| -0.400 | 0.996 | 1.000 | 1.000 | 1.000 | 1.000 | 0.998 |
| -0.600 | 0.997 | 1.000 | 1.000 | 1.000 | 1.000 | 0.999 |
| -0.800 | 0.997 | 1.000 | 1.000 | 1.000 | 1.000 | 0.999 |
| -1.000 | 0.997 | 0.864 | 0.874 | 0.365 | 1.000 | 0.561 |
(iii)Biochemical classes tripeptide Composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| 1.000 | 0.564 | 1.000 | 0.968 | 1.000 | 0.967 | 0.738 |
| 0.800 | 0.721 | 1.000 | 0.980 | 1.000 | 0.979 | 0.840 |
| 0.600 | 0.839 | 1.000 | 0.988 | 1.000 | 0.988 | 0.910 |
| 0.400 | 0.914 | 1.000 | 0.994 | 1.000 | 0.993 | 0.953 |
| 0.200 | 0.946 | 1.000 | 0.996 | 1.000 | 0.996 | 0.971 |
| 0.000 | 0.972 | 1.000 | 0.998 | 1.000 | 0.998 | 0.985 |
| -0.200 | 0.985 | 1.000 | 0.999 | 1.000 | 0.999 | 0.992 |
| -0.400 | 0.993 | 1.000 | 0.999 | 1.000 | 0.999 | 0.996 |
| -0.600 | 0.996 | 1.000 | 1.000 | 1.000 | 1.000 | 0.998 |
| -0.800 | 0.999 | 0.999 | 0.999 | 0.990 | 1.000 | 0.994 |
| -1.000 | 1.000 | 0.680 | 0.703 | 0.196 | 1.000 | 0.365 |
[B] Saccharomyces cerevisiae (i)Amino Acid Composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| 1.000 | 0.263 | 0.971 | 0.617 | 0.900 | 0.568 | 0.331 |
| 0.800 | 0.351 | 0.950 | 0.650 | 0.875 | 0.594 | 0.375 |
| 0.600 | 0.442 | 0.912 | 0.677 | 0.833 | 0.620 | 0.400 |
| 0.400 | 0.527 | 0.855 | 0.691 | 0.784 | 0.644 | 0.405 |
| 0.200 | 0.609 | 0.786 | 0.698 | 0.740 | 0.668 | 0.402 |
| 0.000 | 0.691 | 0.702 | 0.697 | 0.699 | 0.694 | 0.393 |
| -0.200 | 0.762 | 0.600 | 0.681 | 0.656 | 0.716 | 0.367 |
| -0.400 | 0.824 | 0.486 | 0.655 | 0.616 | 0.734 | 0.329 |
| -0.600 | 0.878 | 0.371 | 0.624 | 0.582 | 0.752 | 0.288 |
| -0.800 | 0.920 | 0.264 | 0.592 | 0.556 | 0.768 | 0.245 |
| -1.000 | 0.952 | 0.171 | 0.561 | 0.534 | 0.780 | 0.196 |
(ii)Dipeptide Composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| 1.000 | 0.282 | 0.985 | 0.633 | 0.949 | 0.578 | 0.375 |
| 0.800 | 0.388 | 0.972 | 0.680 | 0.932 | 0.614 | 0.444 |
| 0.600 | 0.489 | 0.948 | 0.718 | 0.903 | 0.650 | 0.491 |
| 0.400 | 0.578 | 0.916 | 0.747 | 0.874 | 0.685 | 0.525 |
| 0.200 | 0.660 | 0.860 | 0.760 | 0.825 | 0.717 | 0.531 |
| 0.000 | 0.728 | 0.786 | 0.757 | 0.773 | 0.743 | 0.515 |
| -0.200 | 0.791 | 0.682 | 0.736 | 0.713 | 0.765 | 0.476 |
| -0.400 | 0.850 | 0.550 | 0.700 | 0.654 | 0.785 | 0.419 |
| -0.600 | 0.900 | 0.404 | 0.652 | 0.602 | 0.802 | 0.351 |
| -0.800 | 0.941 | 0.256 | 0.599 | 0.559 | 0.813 | 0.271 |
| -1.000 | 0.969 | 0.135 | 0.552 | 0.528 | 0.813 | 0.188 |
(iii) Biochemical classes tripeptide Composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| 1.000 | 0.287 | 0.983 | 0.635 | 0.943 | 0.579 | 0.375 |
| 0.800 | 0.385 | 0.968 | 0.676 | 0.923 | 0.611 | 0.434 |
| 0.600 | 0.485 | 0.941 | 0.713 | 0.892 | 0.646 | 0.479 |
| 0.400 | 0.575 | 0.908 | 0.742 | 0.862 | 0.681 | 0.512 |
| 0.200 | 0.656 | 0.852 | 0.754 | 0.816 | 0.713 | 0.519 |
| 0.000 | 0.728 | 0.781 | 0.755 | 0.769 | 0.742 | 0.510 |
| -0.200 | 0.789 | 0.676 | 0.733 | 0.709 | 0.763 | 0.469 |
| -0.400 | 0.846 | 0.550 | 0.698 | 0.653 | 0.782 | 0.416 |
| -0.600 | 0.897 | 0.406 | 0.652 | 0.602 | 0.798 | 0.348 |
| -0.800 | 0.936 | 0.262 | 0.599 | 0.559 | 0.803 | 0.268 |
| -1.000 | 0.962 | 0.144 | 0.553 | 0.529 | 0.792 | 0.185 |
[B] Helicobacter pylori (i)Amino Acid Composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| 1.000 | 0.324 | 0.986 | 0.655 | 0.957 | 0.593 | 0.413 |
| 0.800 | 0.452 | 0.976 | 0.714 | 0.950 | 0.640 | 0.502 |
| 0.600 | 0.559 | 0.954 | 0.757 | 0.924 | 0.684 | 0.558 |
| 0.400 | 0.660 | 0.910 | 0.785 | 0.880 | 0.728 | 0.589 |
| 0.200 | 0.753 | 0.868 | 0.811 | 0.851 | 0.779 | 0.626 |
| 0.000 | 0.825 | 0.800 | 0.813 | 0.805 | 0.821 | 0.626 |
| -0.200 | 0.884 | 0.711 | 0.798 | 0.754 | 0.860 | 0.604 |
| -0.400 | 0.932 | 0.600 | 0.766 | 0.700 | 0.898 | 0.564 |
| -0.600 | 0.958 | 0.476 | 0.717 | 0.646 | 0.919 | 0.496 |
| -0.800 | 0.979 | 0.307 | 0.643 | 0.585 | 0.935 | 0.385 |
| -1.000 | 0.991 | 0.178 | 0.584 | 0.547 | 0.952 | 0.290 |
(ii)Dipeptide Composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| 1.000 | 0.300 | 0.995 | 0.647 | 0.982 | 0.587 | 0.409 |
| 0.800 | 0.486 | 0.988 | 0.737 | 0.975 | 0.658 | 0.547 |
| 0.600 | 0.630 | 0.971 | 0.800 | 0.956 | 0.724 | 0.639 |
| 0.400 | 0.747 | 0.942 | 0.844 | 0.928 | 0.788 | 0.702 |
| 0.200 | 0.824 | 0.910 | 0.867 | 0.902 | 0.838 | 0.737 |
| 0.000 | 0.885 | 0.852 | 0.868 | 0.857 | 0.881 | 0.737 |
| -0.200 | 0.929 | 0.771 | 0.850 | 0.802 | 0.915 | 0.708 |
| -0.400 | 0.965 | 0.661 | 0.813 | 0.740 | 0.950 | 0.657 |
| -0.600 | 0.983 | 0.508 | 0.745 | 0.666 | 0.967 | 0.557 |
| -0.800 | 0.993 | 0.291 | 0.642 | 0.583 | 0.977 | 0.399 |
| -1.000 | 0.997 | 0.121 | 0.559 | 0.531 | 0.973 | 0.244 |
(iii) Biochemical classes tripeptide Composition
| Threshold | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| 1.000 | 0.285 | 0.995 | 0.640 | 0.983 | 0.582 | 0.398 |
| 0.800 | 0.461 | 0.986 | 0.724 | 0.971 | 0.647 | 0.526 |
| 0.600 | 0.630 | 0.970 | 0.800 | 0.954 | 0.724 | 0.637 |
| 0.400 | 0.743 | 0.944 | 0.843 | 0.930 | 0.786 | 0.701 |
| 0.200 | 0.818 | 0.904 | 0.861 | 0.895 | 0.833 | 0.725 |
| 0.000 | 0.877 | 0.846 | 0.862 | 0.851 | 0.873 | 0.724 |
| -0.200 | 0.922 | 0.765 | 0.843 | 0.797 | 0.907 | 0.695 |
| -0.400 | 0.955 | 0.645 | 0.800 | 0.729 | 0.934 | 0.631 |
| -0.600 | 0.980 | 0.490 | 0.735 | 0.658 | 0.961 | 0.540 |
| -0.800 | 0.988 | 0.291 | 0.639 | 0.582 | 0.959 | 0.388 |
| -1.000 | 0.997 | 0.123 | 0.560 | 0.532 | 0.978 | 0.248 |
Following are the methods of feature extraction from amino acid sequence
(i) Amino Acid Composition
This is simply the percentage frequency of all the 20 natural amino acids in the protein sequence. For a single protein sequence the vector represents 20 features.
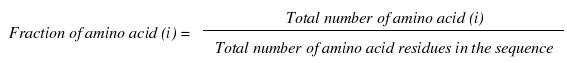
(ii) Dipeptide Composition
This is the percentage frequency of all the 400 dipeptides in the protein sequence. For a single protein sequence the vector represents 400 features.

(iii) Biochemical classes tripeptide Composition
The amino acid residues are classified into six groups based on biochemical similarity. For feature extraction, we consider these 6 classes instead of standard amino acid. So the vector of the sequences has reduced dimensionality compare to vector of the standard sequences. This is the percentage frequency of all the 216 tripeptides in the protein sequence. For a single protein sequence the vector represents 216 features.

mamoon