Home Page of PEP2D
Welcome to Peptide Secondary Structure Prediction server that allows users to predict regular secondary structure in their peptides (e.g., H: Helix, E:Strand, C:Coil). Till date all the secondary structure prediction methods are optimized for proteins. Peptides may adopt diffrent secondary structure when integrated in proteins. Thus it is important to develop seperate method for predicting secondary structure of peptides instead of using protein secondary structure prediction methods.
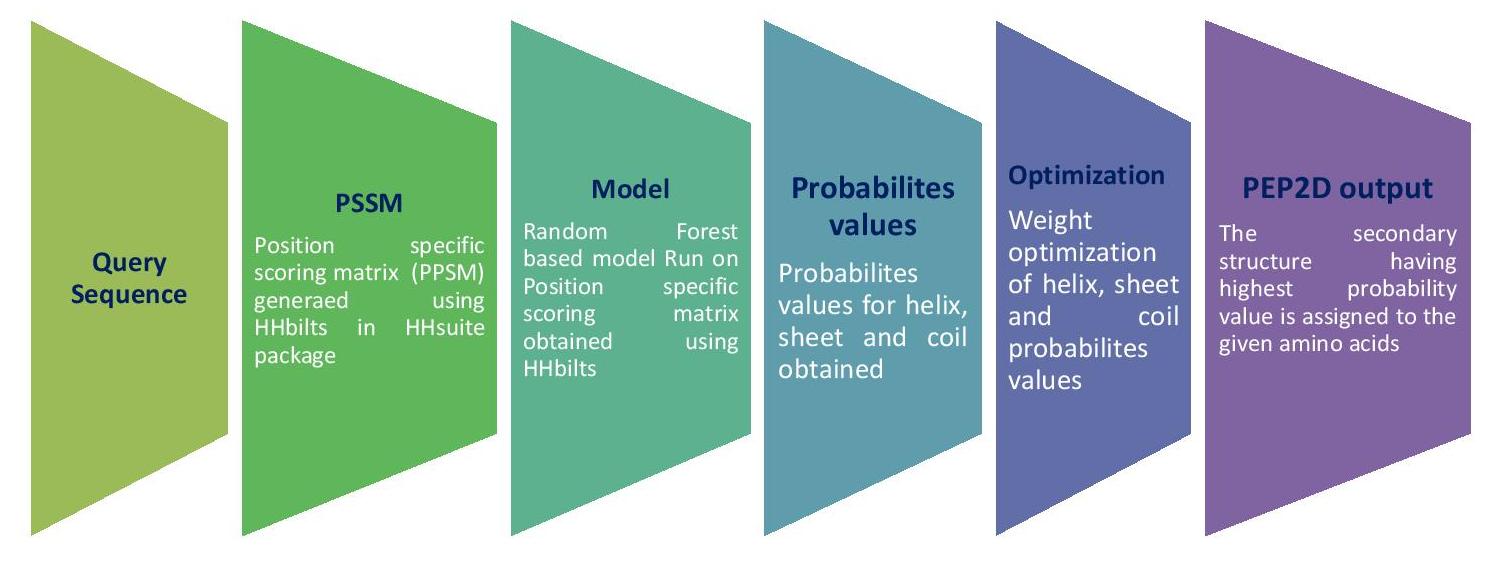
PEP2D server implement models trained and tested on around 3100 peptide structures having number of residues between 5 to 50. This server have following three main modules;
Prediction module: Allow users to predict secondary structure of limitted number of peptides (less than 10 sequences) using PSSM based model (best model).
Multiple Sequences: This module developed for predicting secondary structure multiple peptides (up to 1000 peptide sequences at a time) using binary based models.
Mutant Peptides: This is interesting module developed to design peptide with desired secondary structure. First it generate all possible mutants of a query peptide then it predict secondary structure of each mutant using binary based model.
Reference: Singh H., Singh S. and Raghava G.P.S. (2019) Peptide Secondary Structure Prediction using Evolutionary Information. bioRxiv, 558791; doi.org/10.1101/558791
 PEP2D server implement models trained and tested on around 3100 peptide structures having number of residues between 5 to 50. This server have following three main modules;
PEP2D server implement models trained and tested on around 3100 peptide structures having number of residues between 5 to 50. This server have following three main modules;