MycoPrInt: Algorithm
STEP 1
First of all the Mtb proteome was mapped with Pfam domains with the help of hmmpfam program of HMMER software.
STEP 2
A set of crystallographically verified domain-domain interactions (CVDDIs) have been compiled from 3did database. 3did database uses Pfam domain definition.
STEP 3
The Mtb protein pairs containing at least one CVDDI, such that one domain is present on single protein and the other domain partner is on another protein,were searched and compiled and predicted to be interacting. The figure below shows the schematic diagram of this method.
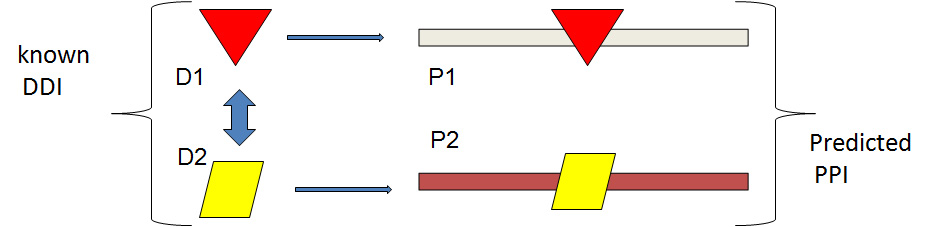
D1D2 (taken from CVDDI set) domain-domain interaction was mapped on protein pair(s). Any two protein (P1P2) containing the D1D2, such that D1 is mapped on P1 and D2 on P2, is predicted as interacting.


