LBtope : Prediction of Linear B-cell EpitopesPredict of B-cell epitopes (antigenic region) with high accuracy is one of the major challenges in designing subunit/peptide vaccine or immunotherapy. In past number of methods have been developed for predicting linear or continious B-cell epitopes like ABCpred. These existing methods have two major limitations, first they developed on small dataset, second random peptides were used as non B-cell epitopes in these methods. 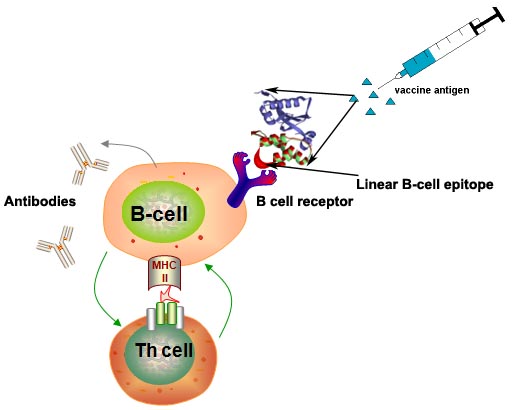 In order to overcome limiations of existing methods, we developed a method LBtope for predicting linear B-cell epitopes. We developed several models using various techniques (e.g., SVM, IBk)on a large dataset of B-cell epitopes and non-epitopes (12063 epitopes and 20589 non epitopes obtained from IEDB database). First time, experimetally valiadted non B-cell epitopes were used for developing prediction model. In order to provide service to the scientific community, we developed a web server LBtope for predicting and designing B-cell epitopes. |
- Singh H, Ansari HR, Raghava GPS (2013) Improved Method for Linear B-Cell
Epitope Prediction Using Antigen's Primary Sequence. PLoS ONE 8(5): e62216 - Singh H, Gupta S, Gautam A, Raghava GP. (2015) Designing B-Cell Epitopes for Immunotherapy and Subunit Vaccines. Methods Mol Biol. 1348:327-40.

