IFNepitope2: Host-specific method for predicting IFN-gamma is avilable
Welcome to IFNepitope Home Page
| This web server is developed for users working in the field of vaccine design. This server allows users to predict and design IFN-gamma inducing peptides. It have following three modules. | ||||||||
| ||||||||
 |
||||||||
|
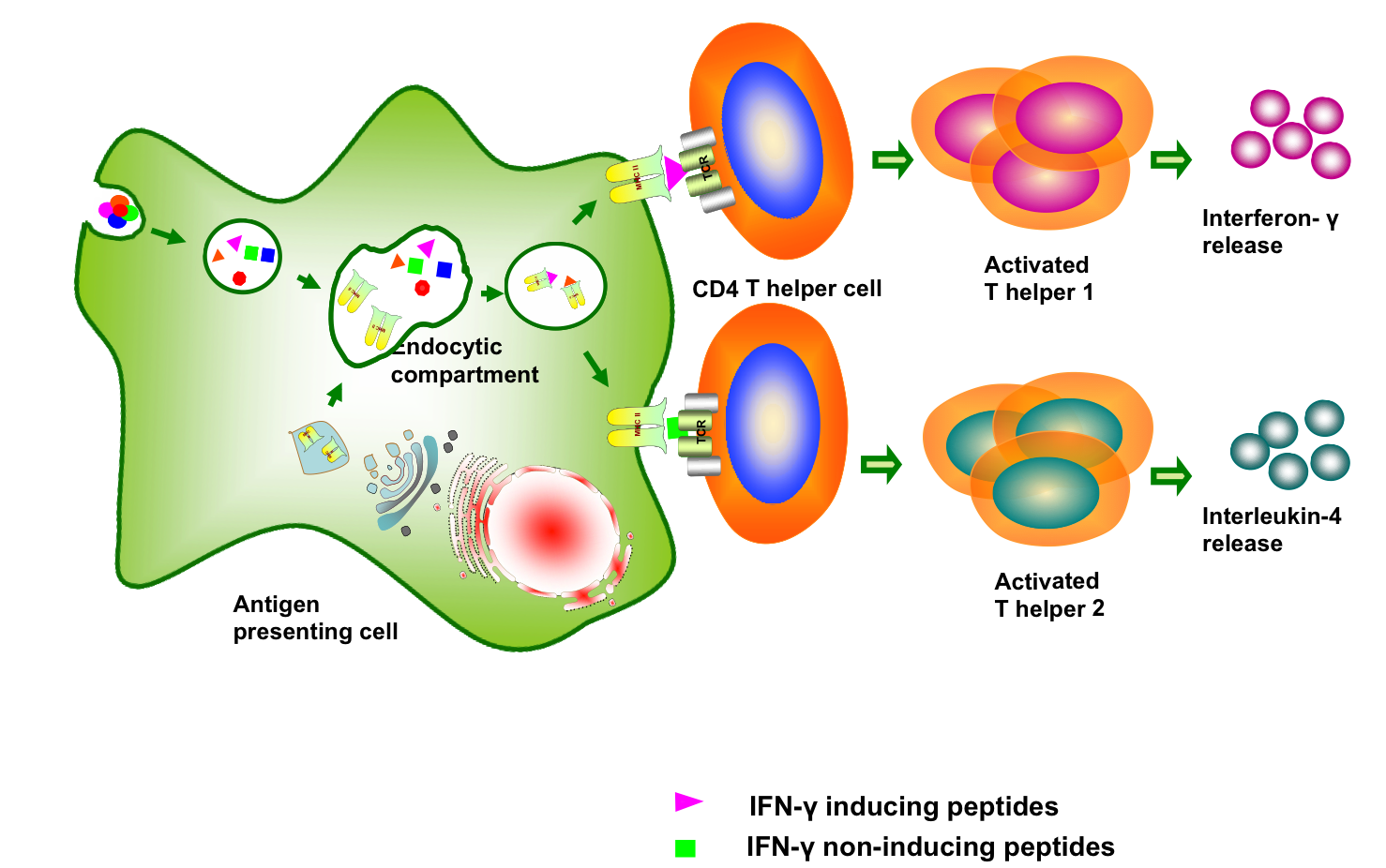
Background:In the modern era, computational biology have paved the way for more systematic approach in designing better and efficacious subunit vaccine against infectious pathogens. With subunit vaccines, the preferred approach is to use a small fragment of the pathogen to trigger specific immune response, rather than presenting whole organism to the host immune system. IFN-gamma is the signature cytokine of both the innate and adaptive immune system eliciting antiviral, immune regulatory as well as anti-tumor activities. The release of IFN-gamma is the major arm of the Th1 response critical for the control of intracellular pathogens, such as Mycobacterium tuberculosis.
IFNepitope is an online prediction server that aims to predict and design the peptides from protein sequences having the capacity to induce IFN-gamma release from CD4+ T cells. This server is developed with the intention to help microbiologists and immunologists to enable them to choose and design IFN-gamma inducing MHC class II binder peptides from proteins of interest to design better and effective subunit vaccines. |
Reference: Dhanda et. al 2013: Designing of interferon-gamma inducing MHC class-II binders. Biology Direct 2013