Welcome to Help Page of FluSPred
This web server has been designed to help the scientific community in vaccine design and development. This server predicts zoonotic hosts of Influenza A virus from Genome or Protein Sequences.
This page provides help on different modules of the server. The web server has two major modules: (i) Genome and (ii) Protein. After selecting either of the models, a single or multiple sequences, of either of the 15 proteins or the genome, can be used as an input in .fasta format, after the corresponding protein model is selected by the user.
When a particular model is selected by the user and clicked on the "Predict" button, FluSPred then runs the respective model and provides the result in the form of a table which can be downloaded in .csv format. An example sequence has been provided in the input section.
The result comprises the prediction of the zoonotic host, whether the particular strain would be capable of crossing the species barrier to infect humans or not.
Genome Prediction
This module allows you to predict zoonotic hosts based on the Genome sequence, to calculate the results based on genome sequences, please see the image for instruction.
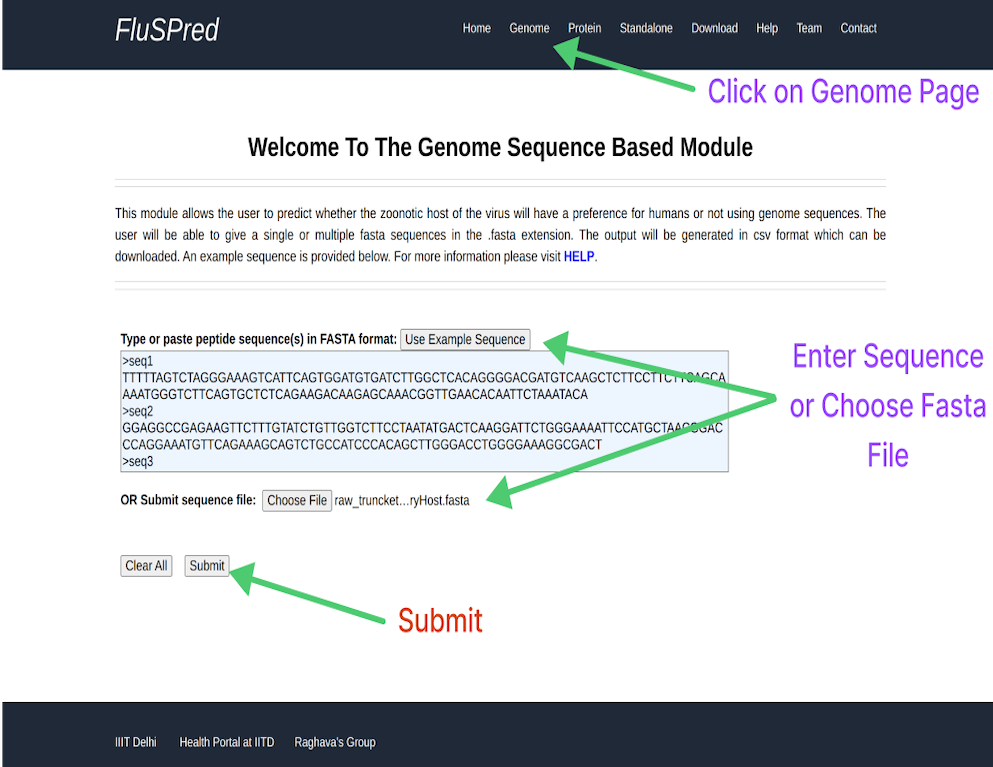
Result
From the result page, you can see the prediction and download the results in a CSV file.

Protein Prediction
This module allows you to predict zoonotic hosts based on the Protein sequences, to calculate the results based on protein sequences, please see the image for instruction. Please choose one of the protein names from the given 15 on the main page.

Result
From the result page, you can see the prediction and download the results in a CSV file.
