Information Page About CancerPPD |
Contents | |
Architecture of CancerPPD |
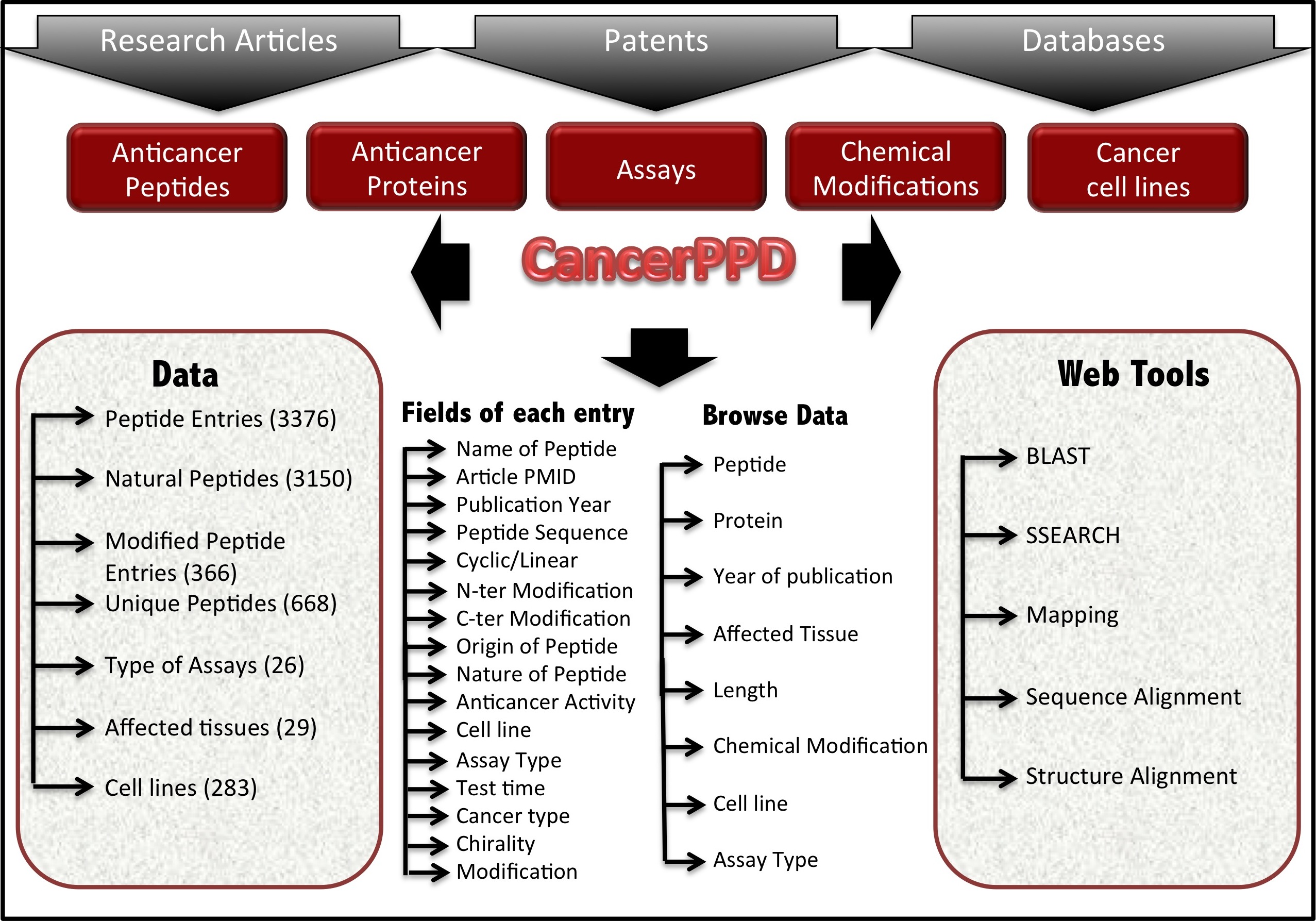 |
Brief description of all the fields in CancerPPD | Go to TOP |
| Name of the Field | Description |
| Name of peptide | This field gives the name of the peptide as mentioned in the article. |
| Article PMID | This provides the PMID of the article from which the entry is made. |
| Publication year | It gives the year in which the article was published. |
| Peptide sequence | This field gives the sequence of the peptide. |
| Cyclic/Linear | This field describes whether the peptide is linear or cyclic |
| N-ter modification | It gives information regarding the N-terminus modification incorporated in the peptide e.g. acetylation |
| C-ter modification | It gives information regarding the C-terminus modification incorporated in the peptide e.g. Amidation |
| Origin of peptide | It describes from where the peptide was extracted e.g. Extracted from skin secretions of frog |
| Nature of peptide | It describes the biological property possessed by the peptide besides anticancer property e.g. antimicrobial |
| Anticancer activity | This field gives the value of anticancer activity shown by the peptide. |
| Cell line | It gives the cell line on which the peptide was tested. |
| Assay type | This field gives name of the assay used to check the anticancer property of the peptide. |
| Test time | This field gives the information about the incubation period. |
| Cancer type | This gives the name of the cancer type for which the peptide was tested against. |
| Chirality | This fields provides the information whether the amino acids of the peptide have D conformation or L conformation. |
| Modification | This field gives the information about the type of chemical modification or non-natural amino acid incorporated in the peptide e.g. Ornithine |
FAQs (Frequently asked questions) | Go to TOP |
|
Q1. What is CancerPPD? Ans. CancerPPD is a manually curated repository of experimentally validated anticancer peptides and proteins. Q2. What are anticancer peptides? Ans. Anticancer peptides are amphipathic, mostly cationic often derived from antimicrobial peptides. As the name suggests, these selectively kill cancerous cells owing to the differences found between the cell membrane of normal and cancerous cells. The proposed mechanism of their action is either cytoplasmic membrane disruption via micellization/pore formation, or induction of apoptosis. Q3. Why CancerPPD Database is created? Ans. In the last decade, small peptides having anticancer properties have emerged as a potential alternative approach for cancer therapy. Peptide-based therapy has numerous advantages over small molecules that involve high specificity, low production cost, high tumor penetration, ease of synthesis and modification etc. Anticancer peptides have benefits over the current anticancer therapies in being more selective, specific and are unaffected by common mechanism of resistance. Keeping in mind their therapeutic importance, we have tried to catalogue all the available updated information on these peptides from literature. Q4. What is unique about CancerPPD? Ans. CancerPPD is a first and unique database of its kind, which provides comprehensive information about anticancer peptides /proteins and the targeted cell lines .It has many online tools. One of the powerful tools is peptide mapping and similarity search, which allows user to search similar peptides/motifs in a query sequence. Also, CancerPPD database provides 3D structures of anticancer peptides. In addition, Cancer PPD links important databases like CCLE and COSMIC to provide the user with complete information about various cancer cell lines. Q5. Does this database represent all the experimentally validated anticancer peptides available in literature? Ans. This database is the result of first round curation and the literature is being continuously searched for more studies with the objective of providing an exhaustive repository. Q6. Why search CancerPPD? Ans. CancerPPD provides all the experimentally validated information about anticancer peptides and proteins such as anticancer activity, nature of peptide, origin of peptide, chirality, amino acid sequence, N- and C- terminal modifications, structure of petide, source of peptide and targeted cell lines. This information may be very useful for researchers working in the area of therapeutic peptides. Apart from this, user can get detailed information about cell lines from linked databases like COSMIC and CCLE. Q7. How do I process a text search with CancerPPD database? Ans. User can search a peptide by name, peptide sequence, assay type, anticancer activity, cell line, year and PMID etc. Q8. Is this database useful if users have their own query sequence? Ans. Yes, user can use tools like BLAST, SMITH-WATERMAN, MUSCLE, peptide mapping, Helical Wheel to obtain information regarding sequence alignment and perform structural studies. Q9. To whom can I report a discrepancy? Ans. Please refer to the "Contact Us" page. |
Statistical information about CancerPPD | Go to TOP | |
 |
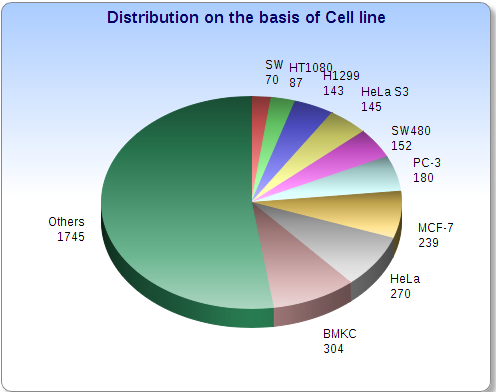 |
 |
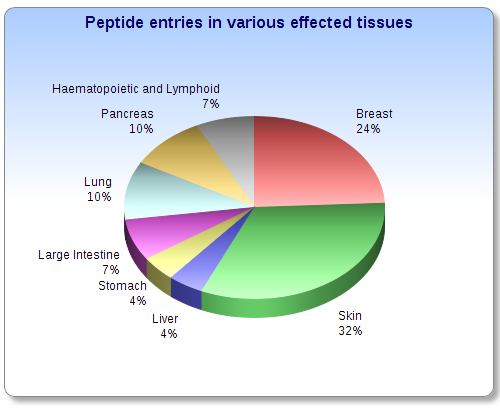 |
