Welcome To The B3Pred Webserver
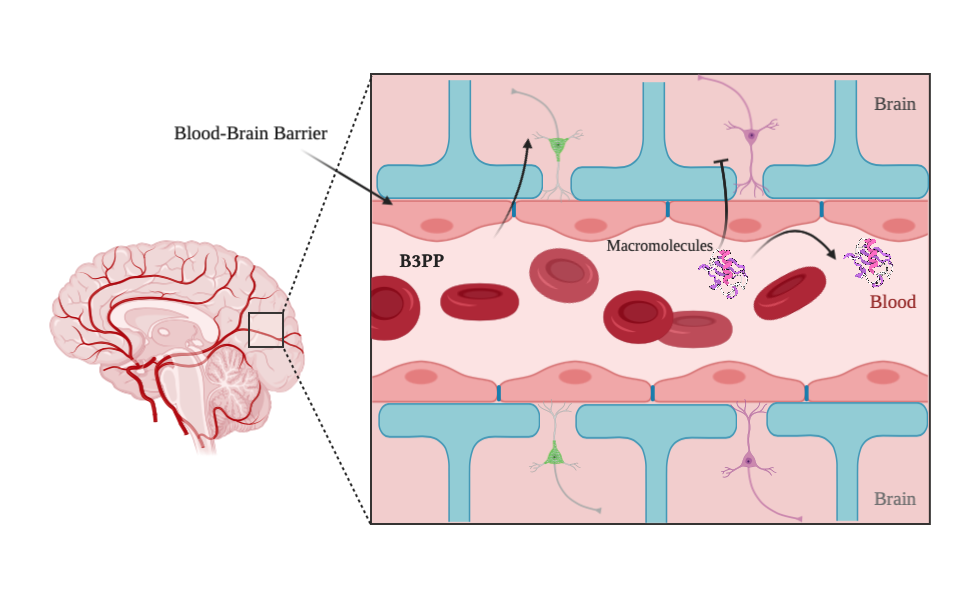
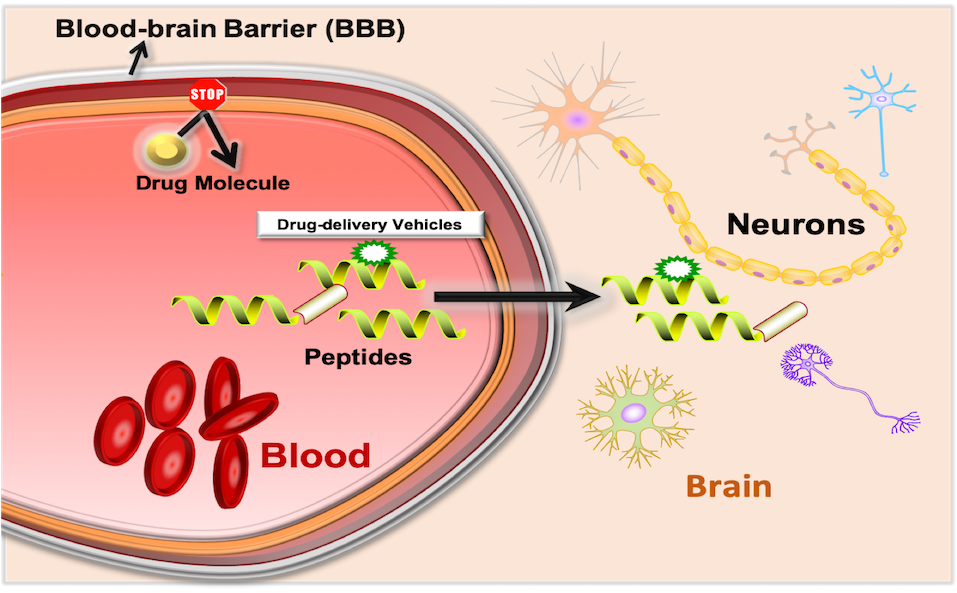
Blood-Brain Barrier Penetrating Peptides Prediction
B3Pred is an in silico method, which is developed to predict and design efficient Blood-Brain Barrier penetrating peptides (B3PPs). The main dataset used in this method consists of 269 experimentally validated B3PPs from B3Pdb database. The dataset prepared for the prediction is following:
* (B3PDB + Random): Dataset comprised of 269 B3PPs (Blood-Brain Barrier Peptides) + 2690 Randomly generated peptides for the negative dataset
Reference: Kumar, V.; Patiyal, S.; Dhall, A.; Sharma, N.; Raghava, G.P.S. B3Pred: A Random-Forest-Based Method for Predicting and Designing Blood–Brain Barrier Penetrating Peptides. Pharmaceutics 2021, 13, 1237.
.
Major Features include:
(1) Peptide Prediction: This module allows user to predict B3PPs on the basis of selected model.
(2) Desing Peptide: This module allows user to generate all possible single mutant analogues of their peptides and predict whether the analogue is B3PPs or not.
(3) Protein Scanning: This module generates all possible overlapping peptides and their single mutant analogues of protein submitted by the user. It also predicts whether overlapping peptide/analogue is B3PP or not.
One of the major features of server is that it also calculates various physicochemical properties. Peptide analogues can be displayed in sorting order based upon desired properties.