|
 |
| | | | | Alpha Turns |
Irregular protein secondary structures are believed to be important structural domains involved in molecular recognition processes and protein folding. In this respect, tight turns are being studied in detail. Depending on the number of residues forming the turn, the tight turns are classified as delta-turns, gamma-turns, beta-turns, alpha-turns and pi-turns(Chou 2000). Among these tight turns, beta- and gamma-turns have been studied in detail and precisely classified. |
In comparison to beta- and gamma-turns, alpha-turns are little investigated due to their lower occurrence in proteins and peptides. The alpha-turn corresponds to a chain reversal involving five amino acids and may be stabilized by a hydrogen bond between the CO group of the first residue and the NH group of the fifth (Pavone et al. 1997). |
In 1997, Pavone et al. had undertaken a systematic search of isolated alpha-turns in a data set of 193 protein X-ray structures.They found 356 alpha turns, of which 238 are of type I-alphaLS, 39 of type II-alphaRS, 8 of type II-alphaLS, 28 of type I-alphaRU, 14 of type I-alphaLU, 9 of type II-alphaRU, 8 of type II-alphaLU, and 7 of type I-alphaC. The categorization of alpha turns into 9 different types is based on the backbone dihedral angles in residues i+1, i+2, and i+3. The representative values of phi(i+1), psi(i+1), phi(i+2), psi(i+2), and phi(i+3), psi(i+3) for each of the nine alpha-turn types as classified by Pavone et al. are given in following Table.
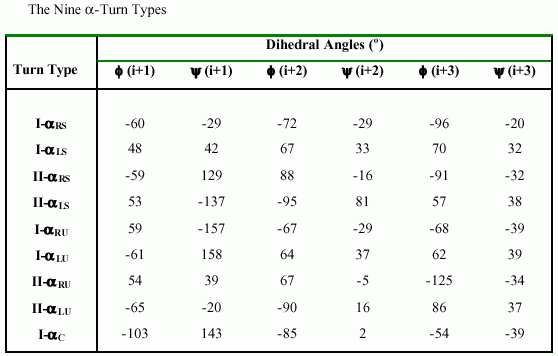
|
Although each of the nine alpha-turn types classified by Pavone et al. has its own special trajectory character, they share some common features; i.e., they all involve five amino acid residues, they all have an intraturn hydrogen bond, and their C(i)-C(i+4) distances are all smaller than 7Å. |
Alpha-turns are mainly characterized by hydrophilic amino acids. It has also been shown that these structures are not only exposed to solvent, but also protrudes outward from the protein surface with a hook-like shape and therefore these structural motifs can function in interaction mechanism (Artymiuk and Blake 1981; Stout CD 1989; Wang et al., 1990). Moreover, alpha-turns are also relevant structural domains in small peptides, particularly in cyclopeptides containing 7-9 residues in their sequence (Zanotti et al., 1989; Iitaka et al., 1974; Di Blasio et al., 1992). |
|