Help page
DrugMint : is a webserver to predict the drug-likeness property of a molecule. This webserver has 3 module: a) Draw Structure b) Virtual Screening c) Analog Designing.
The above three modules were developed to support the variation in the input file provided by the user. In "Draw Structure" user can sketch thier molecule of interest, whereas "Virtual Screening" tool is to be used for screening the a chemical library. The "Analog Design" module is useful for generating the different analogs of a chemical scaffold via using user defined linkers and R-groups.
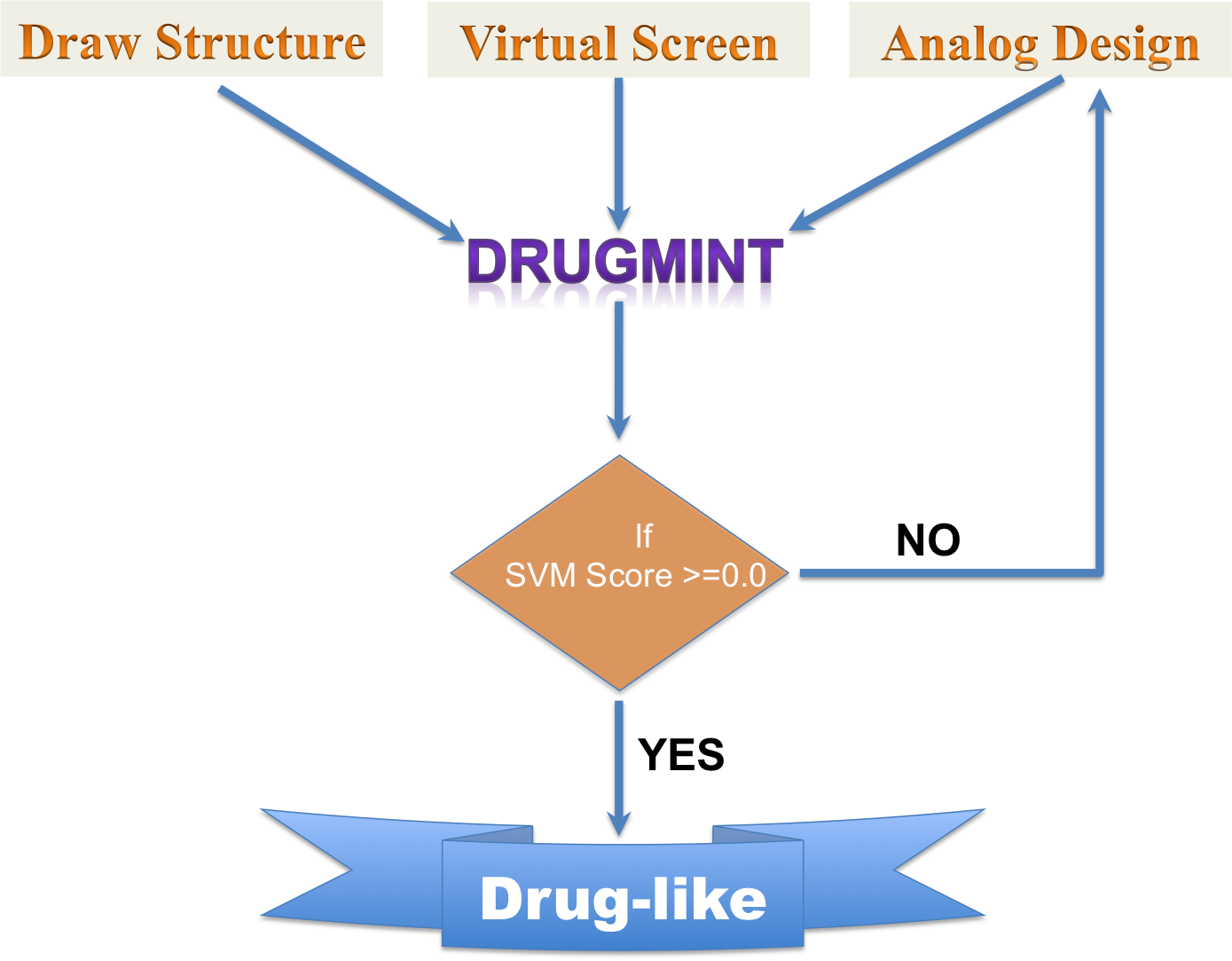
The pictorial representation of the facility in DrugMint
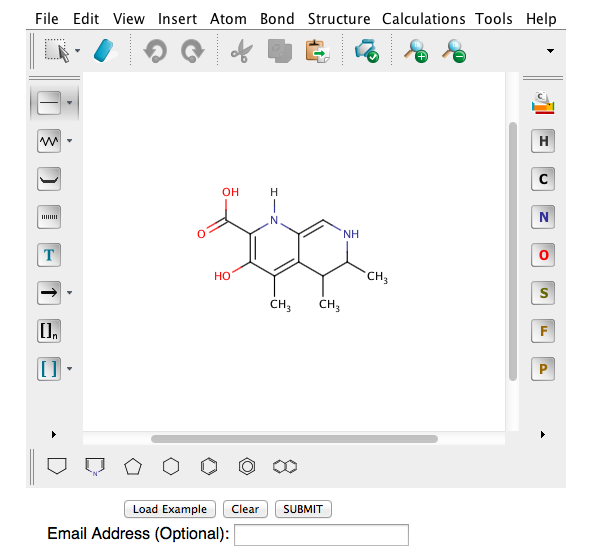
1. INPUT: This server require the structure of chemical as an input. This structure may be in one of the various popular format like sdf, mol, mol2. Here, we have provided three input modules. i) Draw Structure:- This module helps the users to draw the chemcal structure. Using the Marvin applet, users have the choice to either build a new molecule or edit/modify a molecule.
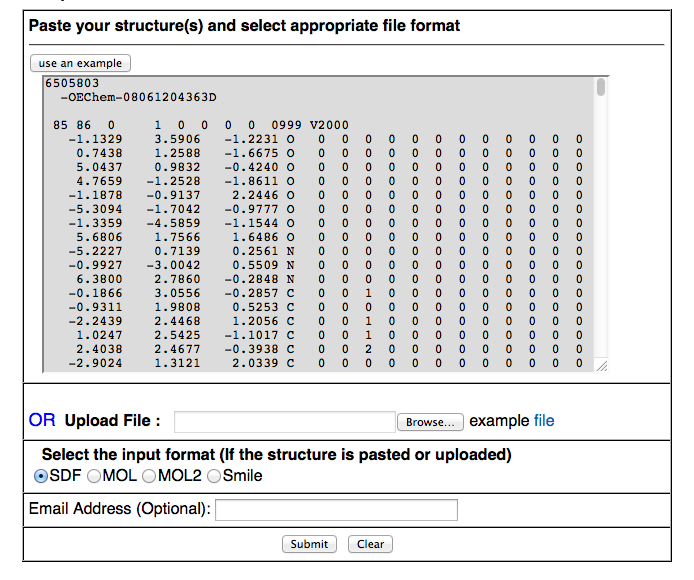
ii) Batch Submission:- Using this tool, users could screen their chemcial library and identify potential drug-like candidates with possibility score to be approved. This module works in two ways 1) Paste molecules with their ids 2) file upload in standard sdf/mol/mol2/smile format.
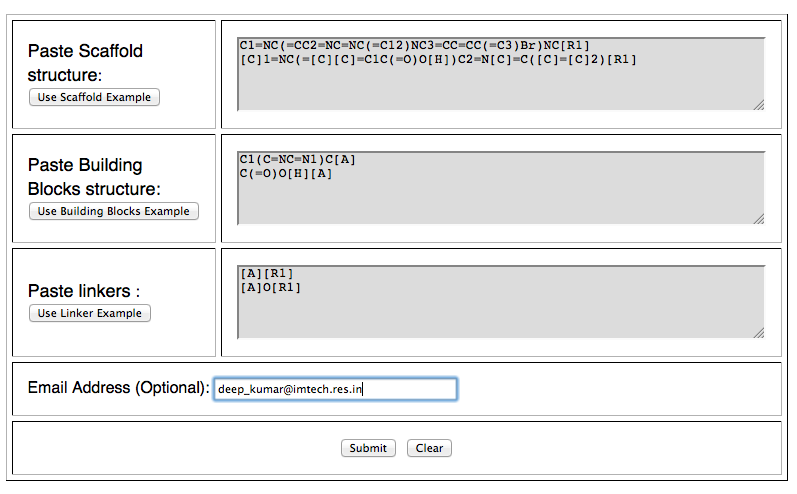
iii) Analog Designing:- To generate new analogs, users will require a scaffold structure, building blocks (R-groups) and linkers. Finally, our server will use the analogs for prediction of drug likeness.
2. PERFORMANCE: The performance of server has been evuluated in five fold cross validation which showed Sensitivity to 77.54%, Specificity 91.65%, Accuracy 87.48% and MCC 0.70%.
3. DATASET: We are also providing the dataset on our dataset link. This dataset comprises of 1348 approved and 3206 experimental drugs as given by Tang K et. al. 2011. One can download from here and can use them for their study.
4. OUTPUT: The output of server will be diplayed in a table that contain drug id, svm score prediction and proabability score. The probability score depicts the confidence score our prediction. This score has been divided in following three categories:
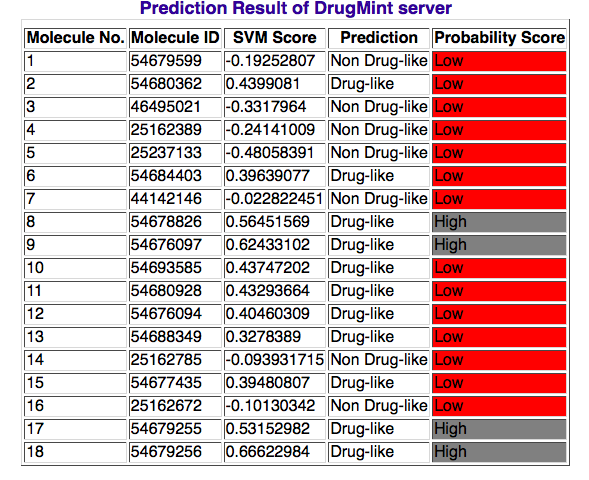
1. Very High : for the svm score greater than or equal to 1.0 (the prediction will be Drug-like and the table row specified for this molecule will be diplayed in background) OR lesser than or equal to -1.0 (the prediction will be Non Drug-like and background color will be default for that particular table row )
2. High : for the svm score between 1.0 and 0.5 (the prediction will be Drug-like and the table row specified for this molecule will be diplayed in background) OR -1.0 and -0.5 (the prediction will be Non Drug-like and background color will be default for that particular table row )
3. Low : for the svm score between 0 and 0.5 (the prediction will be Drug-like and the table row specified for this molecule will be diplayed in background) OR 0.0 and -0.5 (the prediction will be Non Drug-like and background color will be default for that particular table row )

