Algorithm of tRNAmod
In tRNAmod, first we analyzed all the modified tRNA sequences from MODOMICS database. It was observed that most of modifications are uridine-derived, therefore we developed tool for the prediction of uridine-modifications.
Support Vector Machines (SVMs)
SVM is a widely applied and highly successful machine learning technique for the prediction of biological problems. This is based on the structural risk minimization principle of statistics learning theory. We used SVM_light V6.02 package for the development of all prediction models of tRNAmod. It needs complete optimization of different kernels and parameters. There are two software in the package: svm_learn and svm_classify. The svm_learn software used for training and building of a model. After model training, a learned model can be use for the prediction of unknown/test examples using svm_classify.
Nucleotide composition-based approaches
We have applied different nucleotide compositions based approaches such as mono-nucleotide, di-nucleotide and tri-nucleotide compositions for the SVM-based machine learning.
Binary approach
In this approach, first we created overlapping (sliding) window patterns and than these patterns were converted into binary input. We represented A, C, G , U and X with {1,0,0,0,0}, {0,1,0,0,0}, {0,0,1,0,0}, {0,0,0,1,0} and {0,0,0,0,1} respectively.
Structure-based approaches
We applied three different softwares (RNAfold, IPknot and tRNAscan-SE) and used structural information for the prediction modules development.
Hybrid Approach (Binary + tRNAscan-SE)
In the hybrid approaches, we integrated various approahces and found that Hybrid approach of Binary and structural information of tRNAscan-SE performed well together.
Structure visualization of tRNA
We used VARNA software for the structural visualization. VARNA is a Java applet and draws RNA secondary structure. In the result section, structure of all tRNA will be display and prediction highlights with orange color. See example result file
Evaluation Methods
We applied 5-fold cross validation technique for the evaluation of all prediction modules. All predition performances calculated in the terms of Sensitivity, Specificity, Accuracy and MCC using following formulas:
Sensitivity = [TP / (TP+FN)] x 100
Specificity = [TN / (TN+FP)] x 100
Accuracy = [TP+TN / (TP+FP+TN+FN)] x 100
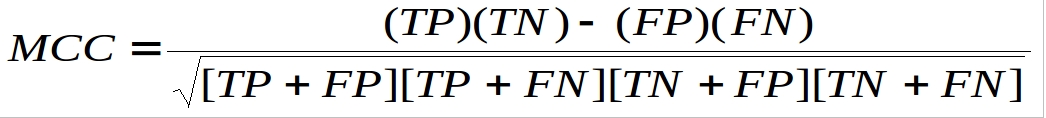
Where TP, TN, FP and FN are True Positive, True Negative, False Positive and False Negative respectively.
Probability Score
The tRNAmod also predicts a probability score for each predicted modified Uridines. It ranges from 0-9 score and calculated by following formula:
Probability score = [(SVM score + 1.5) / 3] x 9
Where maximum and minimum SVM scores rescaled with 1.5 and -1.5 respectively.
