Algortham LBtope
Three different models are implemented in the LBtope | ||
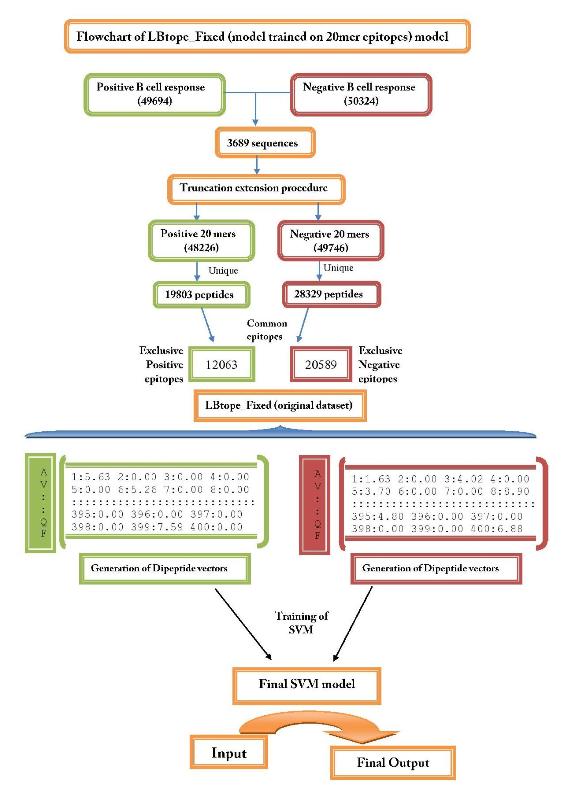
| 1. LBtope_Fixed: This model was developed on LBtope_Fixed dataset, it is suitable for predicting fixed length linear B-cell epitopes of 20 residues. Lbtope_Fixed dataset contain 12063 unique positive patterns and 20589 unique negative patterns, where each pattern having 20-residues. We used extension and termination technique for generating epitopes of length 20 residues. | ||
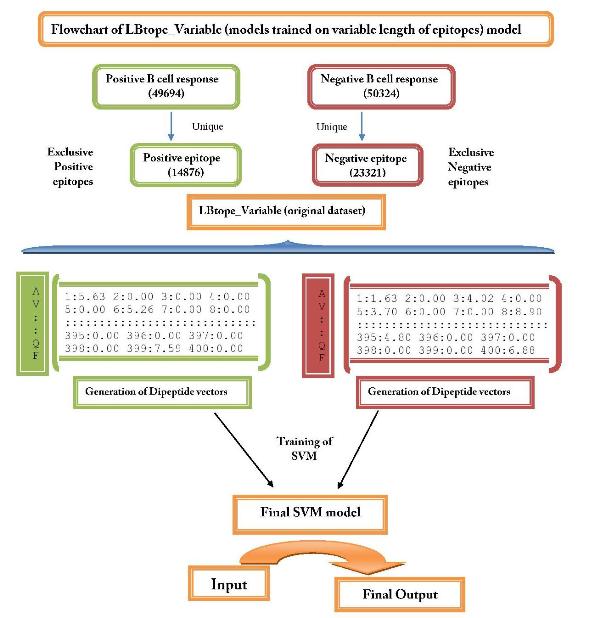
| 2. LBtope_Variable: This model is developed for predicting variable length B-cell epitopes. It is developed on LBtope_Variable dataset that contain 14876 unique B-cell epitopes and 23321 unique non B-cellepitopes. These epitopes and non-epitopes have variable length. | ||
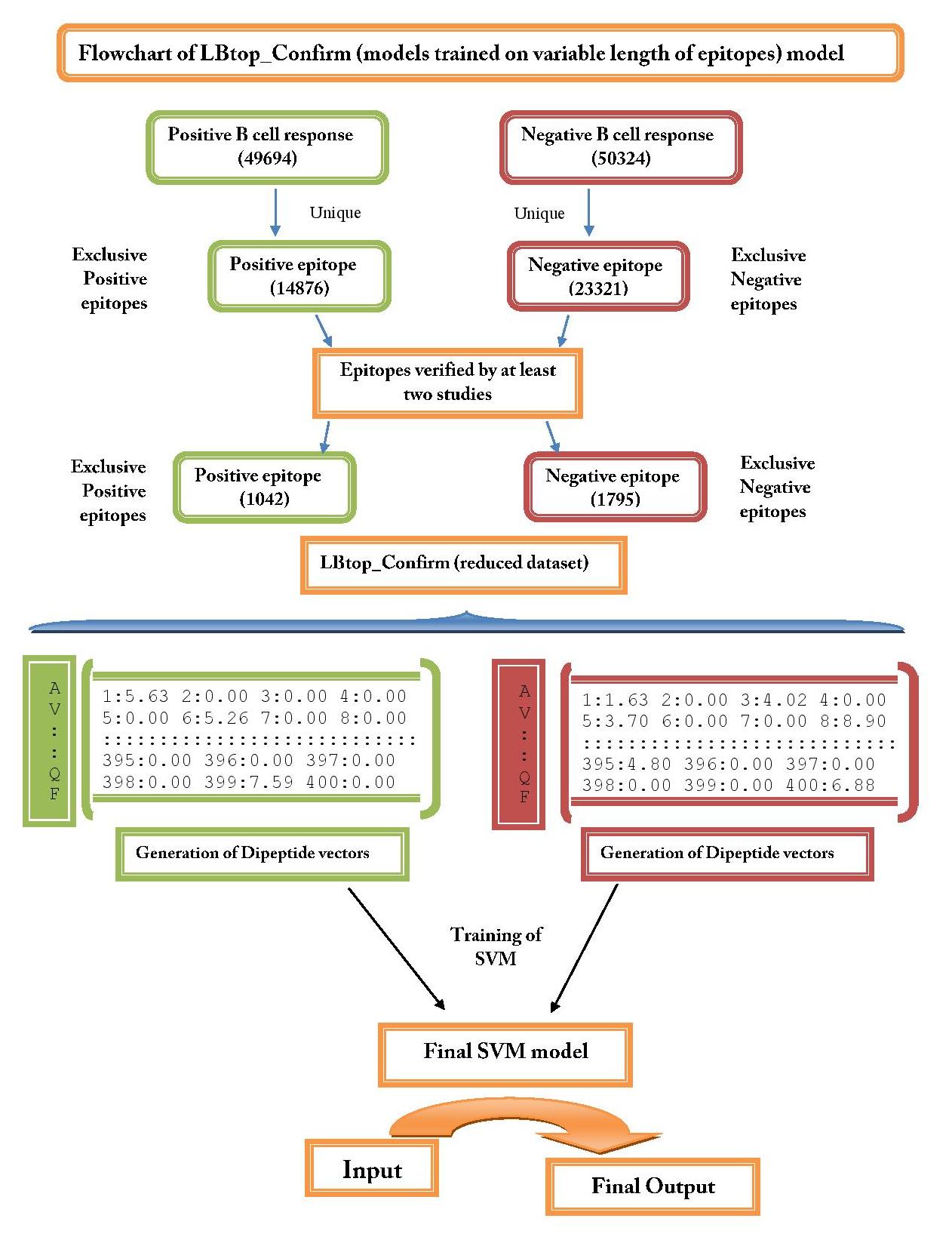
| 3. LBtope_Confirm: This model developed on LBtop_Confirm dataset which contain only those epitopes or non-epitopes that have been experimentally validated by two or more studies. This dataset Lbtope_Confirm contain 1042 unique B-cell epitopes and 1795 unique non B-cell epitopes. | ||
| 4. LBtope_Fixed_non_redundant: This model developed on LBtop_Fixed_non_redundant dataset which is 80% non redundant dataset generated with CD-HIT. LBtope_Fixed_non_redundant dataset contain 7824 unique positive patterns and 7853 unique negative patterns, where each pattern having 20-residues. | ||
| 5. LBtope_Variable_non_redundant: This model developed on LBtop_Variable_non_redundant dataset which is 80% non redundant dataset generated with CD-HIT. LBtop_Variable_non_redundant dataset contain 8011 unique positive patterns and 10868 unique negative patterns, where each pattern having 20-residues. | ||
| 1. LBtope_Fixed | ||
| Go to Top | ||
 | ||
| Go to Top | ||
| 2.LBtope_Variable | ||
| Go to Top | ||
 | ||
| Go to Top | ||
| 3. LBtope_Confirm | ||
| Go to Top | ||
 | ||
| Go to Top |

