|
In past various methods have been develope which predict T-helper epitopes directly or indirectly (MHC Class II binder). Best of our knowledge no method has been developed for predicting epitope or antigenic peptides that can induce IL-10. First time an attempt have been made to develop method for predicting IL-10 inducing epitopes.
The prediction server for IL-10 inducing epitopes has been designed in a very user-friendly manner. Here, on this page, user can get the details of all the algorithms and procedures exploited in the different modules. |
|
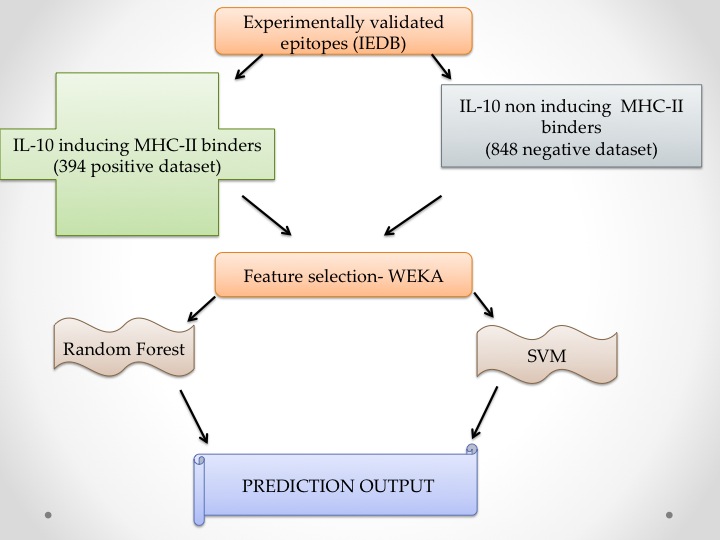
Dataset UsedPositive Dataset:It comprises of 394 MHC class II binders that were reported to cause IL10 release.Negative dataset:It comprises of 848 MHC class II binders which dosenot induces IL-10 secretion. |
This algorithm of the server is relies on the following three models:
Motif based modelAim of this approach is to discover motifs or patterns in IL-10 inducing and non-inducing MHC class II binders. In this study we used a powerful pattern discovery software MERCI. First we identified motifs are exclusively found in IL-10 inducing peptdes and not present in non-inducing peptides. Similarly we identified motifs in non-inducing peptides which are absent in IL-10 inducing peptides. These novel motifs used for discriminating IL-10 inducing and non-inducing peptides.SVM based modelsIn this study, Support Vector Machine (SVM)based models has been developed using software SVM_Light. Major fearures used for training and testing SVM models residue composition of peptides that include amino acid and dipeptide composition. |