| Help |
This page will help users to understand the working of modules available in XIAPin. Table below listed the modules names, user can click them to go directly at any of the mntioned topic.
| Draw & Predict | Screen | Analogs | Potential Inhibitors |
| Draw & Predict |
This tool allow user to draw chemical structure of their molecule by using JME editor. On submission, XIAPin server returns with pIC50 values with an option to calculate chemical descriptors of the query molecule. |
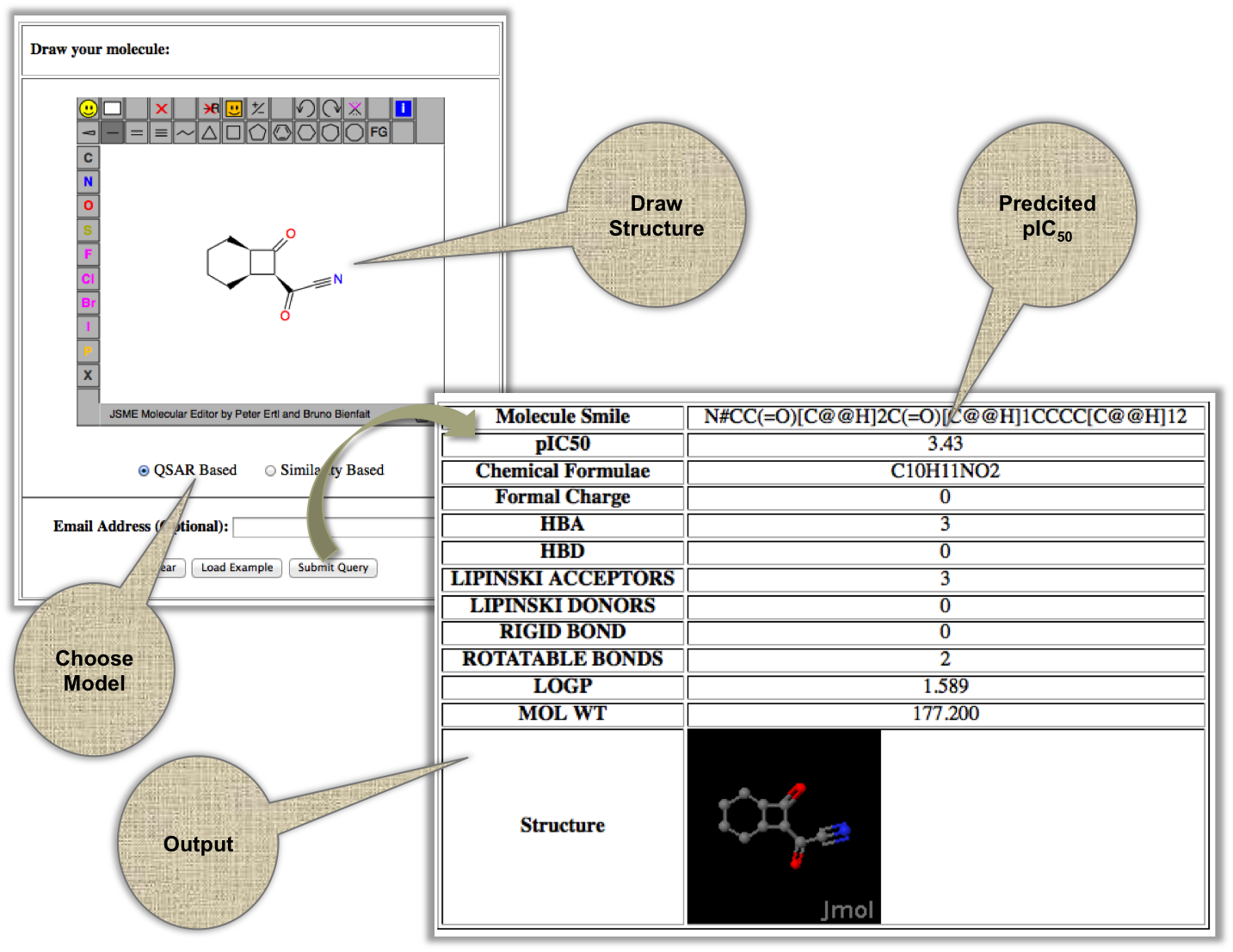 |
| TOP |
| Screen |
In this module, we provide the provision to screen a large number of molecules at one go. User can provide the whole set molecules in SDF format and XIAPin will return their prodicted pIC50. |
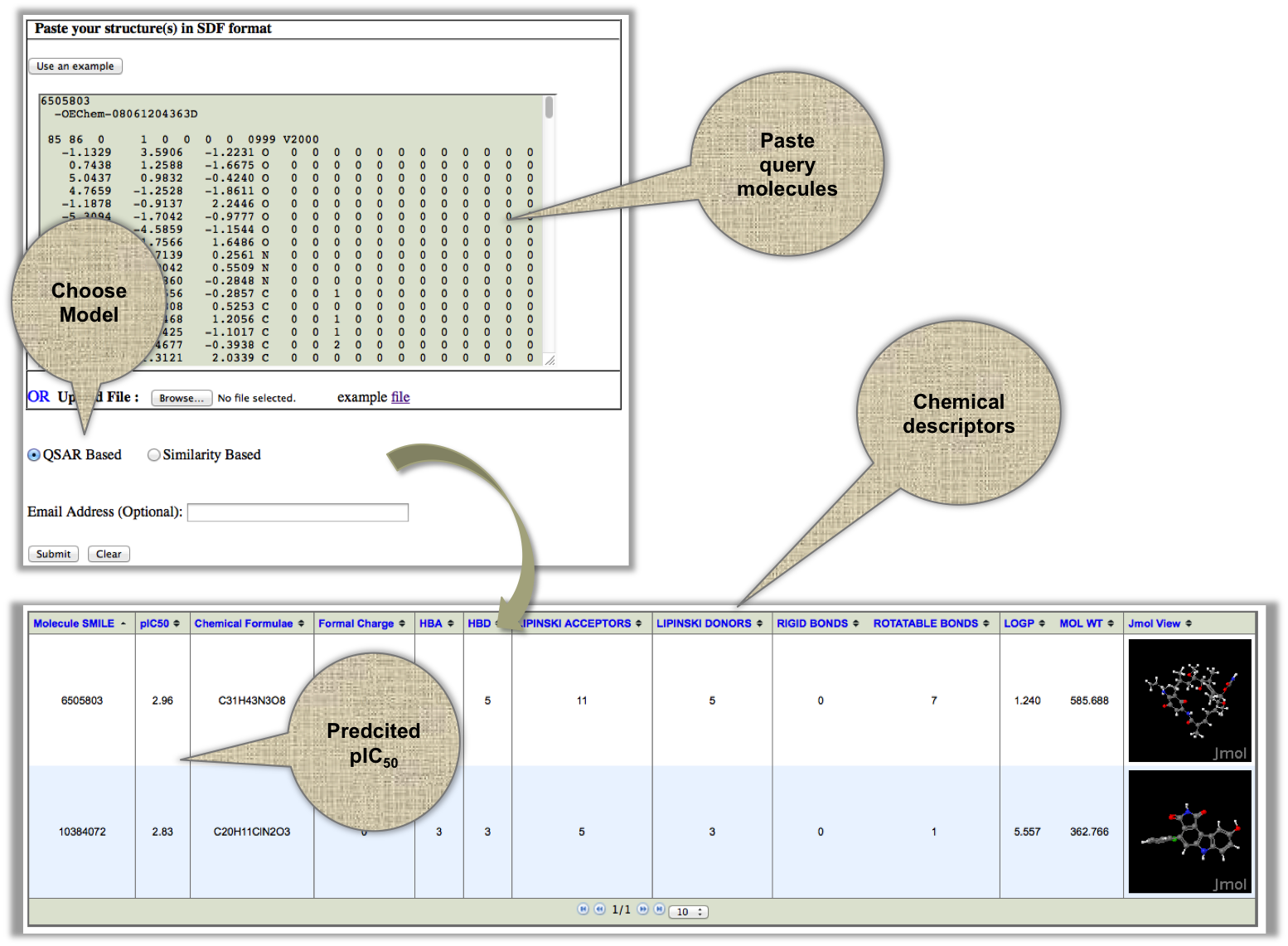 |
| TOP |
| Analogs |
In this module user can design their own molecule by providing the three requirements i.e. scaffold, linkers and building blocks. XIAPin will assemble all these inputs in all the posible way and return all the generated analogs with their pIC50 values. |
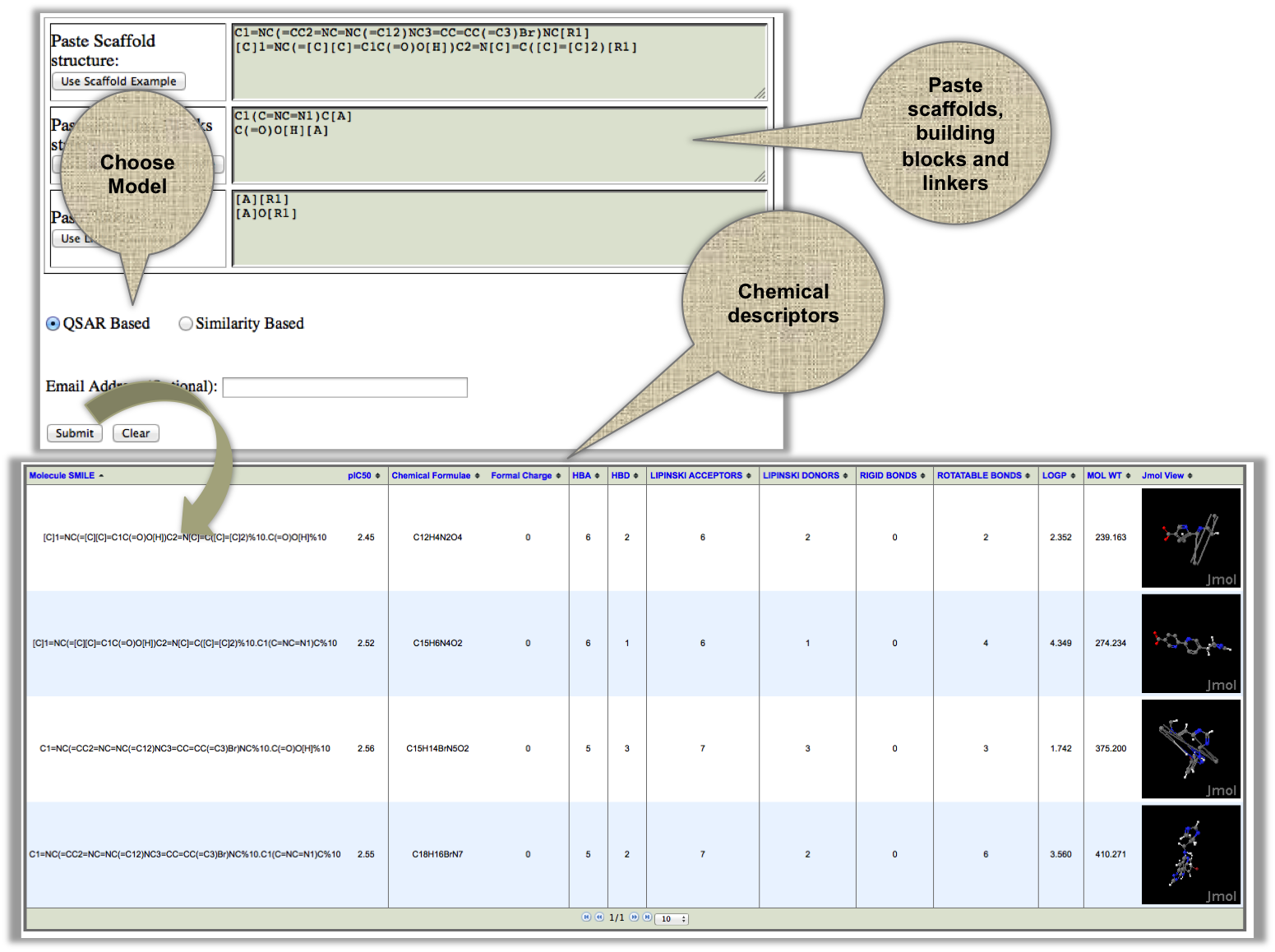 |
| TOP |
| Potential Inhibitors |
In this module, we have provided the list of FDA approved drugs and ZINC molecules with their precomputed pIC50 values, which could be useful from the drug repositioning point of view. |
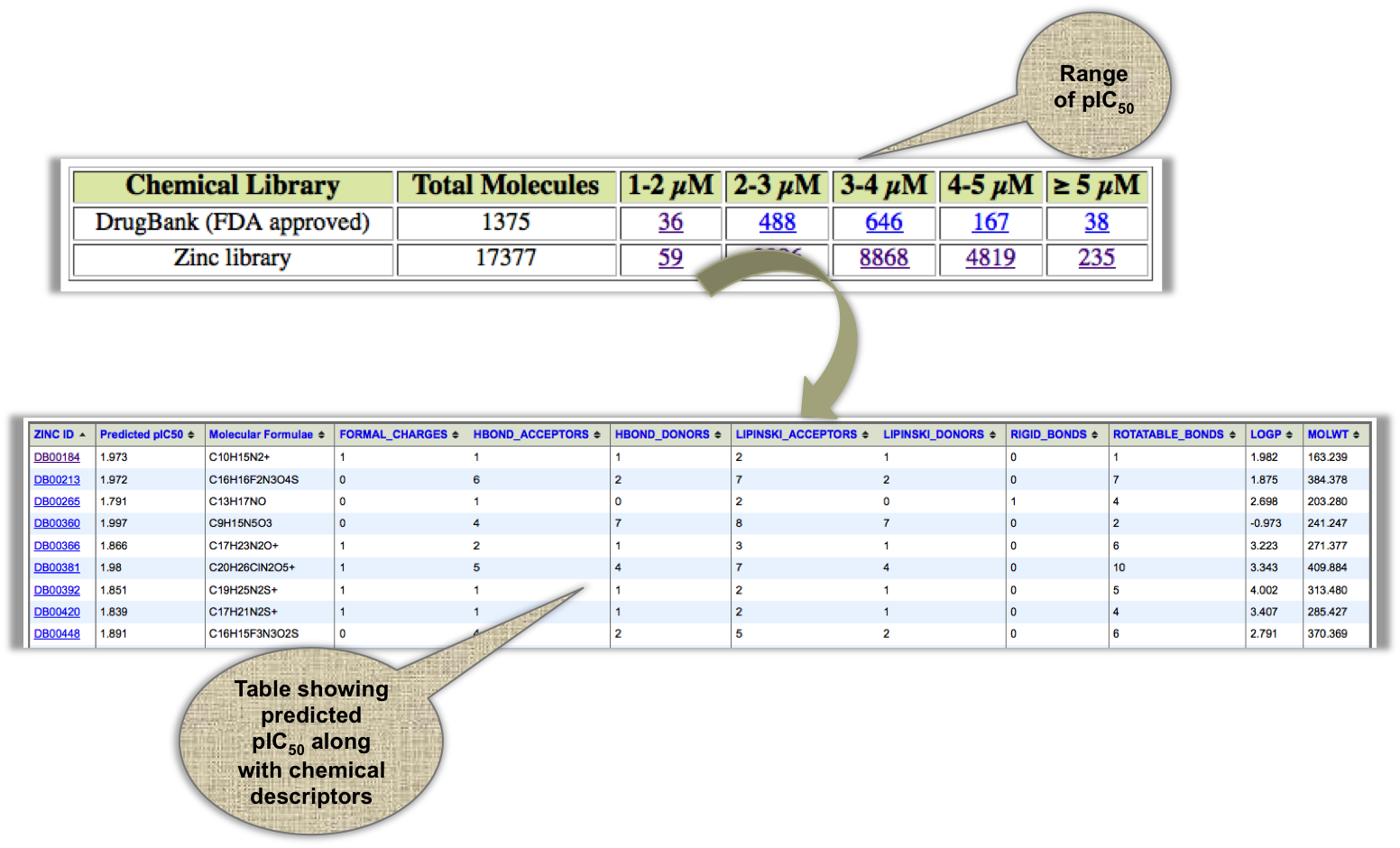 |
| TOP |
