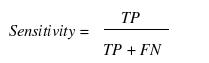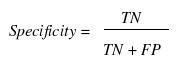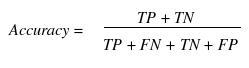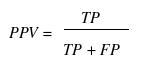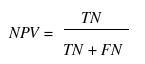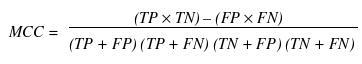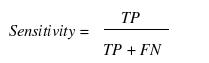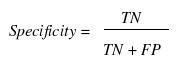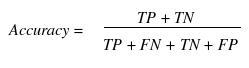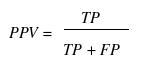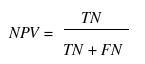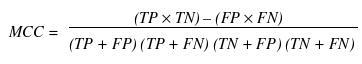ProPrInt: Algorthim
Dataset
Dataset of protein- protein interaction are used separately for each three species namely Escherichia coli, Saccharomyces cerevisiae and Helicobacter pylori. In Escherichia coli dataset, Negative dataset is made from combination of periplasmic and cytoplasmic protein.In Saccharomyces cerevisiae, non-interacting pairs are selected randomly from 4233 yeast proteins. In Helicobacter pylori dataset, if Pairs are not specified explicitly then considered as non-interacting. Datasets are described as follows.
| SI.No | Species | Source | Dataset |
| Interacting | Non Interacting |
| 1 | Escherichia coli | Yellaboina et al., 2007 | 1082 | 13840 |
| 2 | Saccharomyces cerevisiae | Ben-Hur and Noble, 2005 | 10517 | 10517 |
| 3 | Helicobacter pylori | Martin et al., 2005 | 1458 | 1458 |
Approach
Different methods are used to extract the features from the different length of sequences. The methods are Amino Acid composition, Dipeptide composition, Biochemical class tripeptide composition, Pseudo Amino Acid composition and Position specific scoring matrix. Amino Acid composition have 20 features for each protein sequence so totally 40 features for each pair. In this same manner, 800, 232 and 42 features are extracted for Dipeptide composition, Biochemical class tripeptide composition and Pseudo Amino Acid composition
Support Vector Machine
Support Vector Machine is a supervised machine learning methods, used for classifing the protein-protein pair as interacting or non-interacting.SVMlight is an implementation of Support Vector Machines. SVM-light is freely downloadable from this site http://svmlight.joachims.org/. here, we make use of Radial basis function as a kernel function and the results are optimized using the various input parameters. The SVM model was created separately for each species and methods. The training dataset contains both positive data and negative data which are mentioned as 1 and –1 respectively. The model is created from training dataset of known class by using svm_learn and this is used for classifying test dataset using svm_classify.
Performance measure
Various parameters are used for assessing the performance of a method such as specificity, sensitivity, accuracy, Mathew’s correlation coefficient (MCC), positive predictive value (PPV) and negative predictive value (NPV). In order to evaluate this parameter, we calculated true positive (TP), true negative (TN), false positive (FP) and false negative (FN).