| ProGlyProt ID | BC166 |
| Organism Information |
| Organism Name | Pseudomonas aeruginosa 1244 |
| Domain | Bacteria |
| Classification | Family: Pseudomonadaceae
Order: Pseudomonadales
Class: Gammaproteobacteria
Division or phylum: "Proteobacteria" |
| Taxonomic ID (NCBI) | 287 |
| Genome Sequence (s) |
| EMBL | X83916 |
| Gene Information |
| Gene Name | pilA |
| NCBI Gene ID | |
| Protein Information |
| Protein Name | Fimbrial protein (pilin); (group I T4P) |
| UniProtKB/SwissProt ID | P18774 |
| NCBI RefSeq | |
| EMBL-CDS | CAA58768.1 |
| UniProtKB Sequence | >sp|P18774|FM12_PSEAE Fimbrial protein OS=Pseudomonas aeruginosa GN=pilA PE=1 SV=1
MKAQKGFTLIELMIVVAIIGILAAIAIPQYQDYTARTQVTRAVSEVSALKTAAESAILEG
KEIVSSATPKDTQYDIGFTESTLLDGSGKSQIQVTDNQDGTVELVATLGKSSGSAIKGAV
ITVSRKNDGVWNCKITKTPTAWKPNYAPANCPKS
|
| Sequence length | 154 AA |
| Subcellular Location | Surface |
| Function | Structural unit of the pili which have a role in pathogenesis. They are required for colonization through adhesion and and mediating surface translocation (twitching). |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | O (Ser) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | (Propeptide: 1-6) S154 (C-terminal residue) |
| Experimentally Validated Glycosite(s ) in Mature Protein | S148 (C-terminal residue)
|
| Glycosite(s) Annotated Protein Sequence | >sp|P18774|FM12_PSEAE Fimbrial protein OS=Pseudomonas aeruginosa GN=pilA PE=1 SV=1
MKAQKGFTLIELMIVVAIIGILAAIAIPQYQDYTARTQVTRAVSEVSALKTAAESAILEG
KEIVSSATPKDTQYDIGFTESTLLDGSGKSQIQVTDNQDGTVELVATLGKSSGSAIKGAV
ITVSRKNDGVWNCKITKTPTAWKPNYAPANCPKS*(154)
|
| Sequence Around Glycosites (21 AA) | PNYAPANCPKS |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 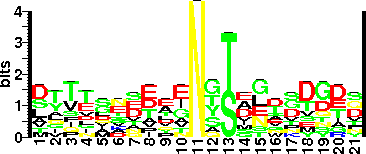 |
| Technique(s) used for Glycosylation Detection | DIG glycan detection (labeling with digoxigenin-succinyl-epsilon-amidocaproic acid hydrazide and anti-DIG antibody after periodate oxidation) |
| Technique(s) used for Glycosylated Residue(s) Detection | N-terminal sequencing and site-directed mutagenesis |
| Protein Glycosylation- Implication | Pilin glycosylation may function to protect the pilus against attack from proteolytic enzymes present as part of the host defense or as produced by P. aeruginosa itself. Presence of glycans, e.g., 5NβOHC47NfmPse on pilin fibrous structure would introduce a negative charge that would lower the pilus isoelectric point, influence solubility, and likely increase ionic interaction among pili and between the pili and extracellular structures therby influencing pilus-dependent functions such as twitch |
| Glycan Information |
| Glycan Annotation | Linkages: β-D-FucNAc-Ser.
A 666.5 Da trisaccharide containing pseudaminic acid, xylose, and N-acetylfucosamine [α-5NβOHC4 7NFmPse-(2→4)-β-Xyl-(1→3)-β-D-FucNAc-(1→3)-β-Ser].
Protein glycan molar ratio is 1:1.
It is structurally identical to the O-antigen repeating unit of P. aeruginosa serotype O7 LPS. This fact together with mutation studies suggest that the metabolic source of the pilin glycan is the O-antigen biosynthetic pathway. The pilin glycan differs from the O- |
| Technique(s) used for Glycan Identification | The gradient NMR spectrum, tandem MS/MS analysis, and methylation analysis provided information on linkage and the sequence of oligosaccharide components. Xylosyl linkage was determined by GLC-MS analysis of partially methylated alditol acetate. |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | pilO (tfpO) |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | PilO (TfpO) |
| OST UniProtKB/ SwissProt ID | |
| OST NCBI RefSeq | |
| OST EMBL-CDS | |
| OST UniProtKB Sequence | |
| OST EC Number (BRENDA) | 2.4.1.119 |
| OST Genome Context | |
| Characterized Accessory Gene(s) | WbpM, WbpL are encoded in the O-antigen biosynthesis cluster. WbpM is a UDP-GlcNAc C6 dehydratase/C4 reductase involved in the UDP-FucNAc biosynthesis. WbpL is a glycosyltransferase that transfers FucNAc from UDP-FucNAc to undecaprenol phosphate. |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Horzempa, J., Dean, C.R., Goldberg, J.B. and Castric, P. (2006) Pseudomonas aeruginosa 1244 pilin glycosylation: glycan substrate recognition. J Bacteriol, 188, 4244-4252. [PubMed: 16740931]
2) Smedley, J.G., 3rd, Jewell, E., Roguskie, J., Horzempa, J., Syboldt, A., Stolz, D.B. and Castric, P. (2005) Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect Immun, 73, 7922-7931. [PubMed: 16299283]
3) Comer, J.E., Marshall, M.A., Blanch, V.J., Deal, C.D. |
| Additional Comments | O-antigen sugars are not sequentially added to the pilin. wbpM and wbpL are essential for the initial steps of O-antigen biosynthesis.
5-N-3 hydroxybutyryl-7-N-formylpseudaminic acid is a part of a trisaccharide modification on P. aeruginosa pilin. It is the second example of glycoprotein with pseudaminic acid containing glycan apart from Campylobacter flagellin protein.
The substrate recognition features required for catalysis by PilO enzyme have been shown to be present in the first |
| Year of Identification | 1995 |
| Year of Validation | 2002 |


