| ProGlyProt ID | BC164 |
| Organism Information |
| Organism Name | Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / NCIB 9290) |
| Domain | Bacteria |
| Classification | Family: Sphingobacteriaceae
Order: "Sphingobacteriales"
Class: "Sphingobacteria"
Division or phylum: "Bacteroidetes" |
| Taxonomic ID (NCBI) | 485917 |
| Genome Sequence (s) |
| GeneBank | CP001681.1 |
| EMBL | CP001681 |
| Gene Information |
| Gene Name | cslB (Phep_0789) |
| NCBI Gene ID | 8251878 |
| GenBank Gene Sequence | 8251878 |
| Protein Information |
| Protein Name | Chondroitinase-B |
| UniProtKB/SwissProt ID | Q46079 |
| NCBI RefSeq | YP_003091073.1 |
| EMBL-CDS | ACU03011.1 |
| UniProtKB Sequence | >sp|Q46079|CSLB_PEDHD Chondroitinase-B OS=Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / NCIB 9290) GN=cslB PE=1 SV=2
MKMLNKLAGYLLPIMVLLNVAPCLGQVVASNETLYQVVKEVKPGGLVQIADGTYKDVQLI
VSNSGKSGLPITIKALNPGKVFFTGDAKVELRGEHLILEGIWFKDGNRAIQAWKSHGPGL
VAIYGSYNRITACVFDCFDEANSAYITTSLTEDGKVPQHCRIDHCSFTDKITFDQVINLN
NTARAIKDGSVGGPAMYHRVDHCFFSNPQKPGNAGGGIRIGYYRNDIGRCLVDSNLFMRQ
DSEAEIITSKSQENVYYGNTYLNCQGTMNFRHGDHQVAINNFYIGNDQRFGYGGMFVWGS
RHVIACNYFELSETIKSRGNAALYLNPGAMASEHALAFDMLIANNAFINVNGYAIHFNPL
DERRKEYCAANRLKFETPHQLMLKGNLFFKDKPYVYPFFKDDYFIAGKNSWTGNVALGVE
KGIPVNISANRSAYKPVKIKDIQPIEGIALDLNALISKGITGKPLSWDEVRPYWLKEMPG
TYALTARLSADRAAKFKAVIKRNKEH
|
| Sequence length | 506 AA |
| Subcellular Location | Periplasm |
| Function | GAG lyase. Cleaves the glycosaminoglycan, dermatan sulfate. EC= 4.2.2.19. |
| Protein Structure |
| PDB ID | 1DBG, 1DBO |
| Glycosylation Status |
| Glycosylation Type | O (Ser) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | S234 |
| Experimentally Validated Glycosite(s ) in Mature Protein | S234 |
| Glycosite(s) Annotated Protein Sequence | >sp|Q46079|CSLB_PEDHD Chondroitinase-B OS=Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / NCIB 9290) GN=cslB PE=1 SV=2
MKMLNKLAGYLLPIMVLLNVAPCLGQVVASNETLYQVVKEVKPGGLVQIADGTYKDVQLI
VSNSGKSGLPITIKALNPGKVFFTGDAKVELRGEHLILEGIWFKDGNRAIQAWKSHGPGL
VAIYGSYNRITACVFDCFDEANSAYITTSLTEDGKVPQHCRIDHCSFTDKITFDQVINLN
NTARAIKDGSVGGPAMYHRVDHCFFSNPQKPGNAGGGIRIGYYRNDIGRCLVDS*(234)NLFMRQ
DSEAEIITSKSQENVYYGNTYLNCQGTMNFRHGDHQVAINNFYIGNDQRFGYGGMFVWGS
RHVIACNYFELSETIKSRGNAALYLNPGAMASEHALAFDMLIANNAFINVNGYAIHFNPL
DERRKEYCAANRLKFETPHQLMLKGNLFFKDKPYVYPFFKDDYFIAGKNSWTGNVALGVE
KGIPVNISANRSAYKPVKIKDIQPIEGIALDLNALISKGITGKPLSWDEVRPYWLKEMPG
TYALTARLSADRAAKFKAVIKRNKEH
|
| Sequence Around Glycosites (21 AA) | RNDIGRCLVDSNLFMRQDSEA |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 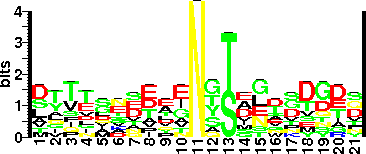 |
| Technique(s) used for Glycosylation Detection | Deduced Crystal structure |
| Technique(s) used for Glycosylated Residue(s) Detection | Crystallographic analysis (electron density maps) |
| Protein Glycosylation- Implication | |
| Glycan Information |
| Glycan Annotation | Linkages: Man- Ser.
The approx. weight of attached glycan was 1180 Da.
Branched heptasaccharide is: galactose-β(1–4)[galactose-α(1–3)](2-O-Me)fucose-β(1–4)xylose-β (1–4)glucuronic acid-α(1–2)[rhamnose-α(1–4)]mannose-α(1-O)Ser. |
| Technique(s) used for Glycan Identification | Crystallographic analysis (electron density maps) |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | |
| OST UniProtKB/ SwissProt ID | |
| OST NCBI RefSeq | |
| OST EMBL-CDS | |
| OST UniProtKB Sequence | |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | Information currently not available with us. |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Michel, G., Pojasek, K., Li, Y., Sulea, T., Linhardt, R.J., Raman, R., Prabhakar, V., Sasisekharan, R. and Cygler, M. (2004) The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a calcium-dependent catalytic machinery. J Biol Chem, 279, 32882-32896. [PubMed: 15155751]
2) Huang, W., Matte, A., Li, Y., Kim, Y.S., Linhardt, R.J., Su, H. and Cygler, M. (1999) Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a |
| Additional Comments | Sequon features: Identified glycosylation Sequon features: Consensus sequon observed Asp-Ser or Asp-Thr-Thr, at turns. |
| Year of Identification | 1999 |
| Year of Validation | 1999 |


