| ProGlyProt ID | BC146 |
| Organism Information |
| Organism Name | Geobacillus tepidama |
| Domain | Bacteria |
| Classification | Family: Bacillaceae
Order: Bacillales
Class: Bacilli (or Firmibacteria)
Division or phylum: "Firmicutes" |
| Taxonomic ID (NCBI) | 265948 |
| Genome Sequence (s) |
| EMBL | AY883421 |
| Gene Information |
| Gene Name | sgtA |
| NCBI Gene ID | |
| Protein Information |
| Protein Name | SgtA |
| UniProtKB/SwissProt ID | Q58R21 |
| NCBI RefSeq | |
| EMBL-CDS | AAX46285.1 |
| UniProtKB Sequence | >tr|Q58R21|Q58R21_9BACI SgtA OS=Geobacillus tepidamans GN=sgtA PE=4 SV=1
MSKKKAIKLATASAIAASAFVAANPNASEAATNVDAVVSQAKAQMRNAYYTYSHTVTNTG
EFADIKAVYAEYNKAKKAYADAVALVKKAGGANKDAYLADLEATYEVYITSRVIPYIDAY
NYAVKLDAMRQELQAAVDAKDLAKVEELYHKISYELKSRTVILDRVYGQTTRDLLRSQFK
AAAQQLRDSLIYDVTVSMKLKAAEEATNKGDLAAAEKALAEAQDALTKATAFKADLTTKQ
ATVKAAYEAKVAPAVVGVSAVNSRTVQIKFNKPIDASTVLVDKGTTDTSDDTLKTTSIVF
TALDGQPAISQTDIKASLSDDKKTLTLVAANSDFFTKRYVVDIKNVKTTDGKDVPAYTTT
IDTTDSVRPAVLSYSYADNGLTLKVKFSEPLNSVGTVKLYDGTTEISVSPSFTAGSDEMT
INLASSSVPVNKALTLKIFGAVDYNGNVINPNPAELTVMKTTVDTTKPTVQSVEAVNDKT
VKVTFSEKLLGNPTIKIGGTTAASVSVDSTGLVYTATLNSAQLGIQAVEVSSYTDLAGNV
GDAYTKVVELKADTTAPKLVSSQVVKINGIENLVLTFDEEVTTQNNITVVSSNDYYVDEN
NVRKQVGVTLTTNSTNFKLYNPVNGKSKSVALDISSLPKGTYTVTLPTGATALVKDLASN
PNAYAEGKQITFVRGTDSLTTKPALDTNVSTNGVEVVDNNTLRFTFTQNLDASALNLSNF
NINGVALSKAVFDGATNKILVTLAPGANTWTGTHVITVSNIKNVSGIAMDTVTTTESMNE
NVAPTFTATLTSADVIKVTFSEPVAHSTLSNAITGNNFTVKVDGTLNNVTGVYTDSAATT
PVSATGNTGYKTVYLKLATPVTDLTKPITVSATNLVDVSAEGASPTVVGNTVSSDVVNVA
K
|
| Sequence length | 901 AA |
| Subcellular Location | Surface |
| Function | Surface layer glycoprotein |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | O (Ser) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | S792 |
| Experimentally Validated Glycosite(s ) in Mature Protein | S792 |
| Glycosite(s) Annotated Protein Sequence | >tr|Q58R21|Q58R21_9BACI SgtA OS=Geobacillus tepidamans GN=sgtA PE=4 SV=1
MSKKKAIKLATASAIAASAFVAANPNASEAATNVDAVVSQAKAQMRNAYYTYSHTVTNTG
EFADIKAVYAEYNKAKKAYADAVALVKKAGGANKDAYLADLEATYEVYITSRVIPYIDAY
NYAVKLDAMRQELQAAVDAKDLAKVEELYHKISYELKSRTVILDRVYGQTTRDLLRSQFK
AAAQQLRDSLIYDVTVSMKLKAAEEATNKGDLAAAEKALAEAQDALTKATAFKADLTTKQ
ATVKAAYEAKVAPAVVGVSAVNSRTVQIKFNKPIDASTVLVDKGTTDTSDDTLKTTSIVF
TALDGQPAISQTDIKASLSDDKKTLTLVAANSDFFTKRYVVDIKNVKTTDGKDVPAYTTT
IDTTDSVRPAVLSYSYADNGLTLKVKFSEPLNSVGTVKLYDGTTEISVSPSFTAGSDEMT
INLASSSVPVNKALTLKIFGAVDYNGNVINPNPAELTVMKTTVDTTKPTVQSVEAVNDKT
VKVTFSEKLLGNPTIKIGGTTAASVSVDSTGLVYTATLNSAQLGIQAVEVSSYTDLAGNV
GDAYTKVVELKADTTAPKLVSSQVVKINGIENLVLTFDEEVTTQNNITVVSSNDYYVDEN
NVRKQVGVTLTTNSTNFKLYNPVNGKSKSVALDISSLPKGTYTVTLPTGATALVKDLASN
PNAYAEGKQITFVRGTDSLTTKPALDTNVSTNGVEVVDNNTLRFTFTQNLDASALNLSNF
NINGVALSKAVFDGATNKILVTLAPGANTWTGTHVITVSNIKNVSGIAMDTVTTTESMNE
NVAPTFTATLTS*(792)ADVIKVTFSEPVAHSTLSNAITGNNFTVKVDGTLNNVTGVYTDSAATT
PVSATGNTGYKTVYLKLATPVTDLTKPITVSATNLVDVSAEGASPTVVGNTVSSDVVNVA
K
|
| Sequence Around Glycosites (21 AA) | VAPTFTATLTSADVIKVTFSE |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 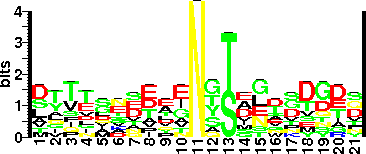 |
| Technique(s) used for Glycosylation Detection | Periodic acid-Schiff (PAS) staining and chemical deglycosylation |
| Technique(s) used for Glycosylated Residue(s) Detection | ESI-QTOF-MS (electrospray ionization quadrupole-time-of-flight mass spectrometry) |
| Protein Glycosylation- Implication | |
| Glycan Information |
| Glycan Annotation | Linkage: β-D-Gal-Ser/Thr.
[→2)-α-L-Rhap-(1→3)-α-D-Fucp-(1→]n=~20. Rhamnose residue at the nonreducing end of the terminal repeating unit of the glycan chain is di-substituted. (R)-N-acetylmuramic acid found in an α-linkage to carbon 3 of the terminal rhamnose residue, serving as capping motif of an S-layer glycan. In addition, that rhamnose is substituted at position 2 with a β-N-acetylglucosamine residue. The S-layer glycan chains are bound via the trisaccharide core →2)-α-L- |
| Technique(s) used for Glycan Identification | HPAEC/PAD (high performance anion-exchange chromatography/pulsed amperometric detection), 1H and 13C NMR spectroscopy including 2D NOESY (nuclear Overhauser enhancement spectroscopy), HMBC (heteronuclear multiple bond correlation spectroscopy) and TOCSY (total correlation spectroscopy) |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | |
| OST UniProtKB/ SwissProt ID | |
| OST NCBI RefSeq | |
| OST EMBL-CDS | |
| OST UniProtKB Sequence | |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Messner, P., Steiner, K., Zarschler, K. and Schaffer, C. (2008) S-layer nanoglycobiology of bacteria. Carbohydr Res, 343, 1934-1951. [PubMed: 18336801]
2) Zayni, S., Steiner, K., Pfostl, A., Hofinger, A., Kosma, P., Schaffer, C. and Messner, P. (2007) The dTDP-4-dehydro-6-deoxyglucose reductase encoding fcd gene is part of the surface layer glycoprotein glycosylation gene cluster of Geobacillus tepidamans GS5-97T. Glycobiology, 17, 433-443. [PubMed: 17202151]
3) Kahlig, H., Kolarich |
| Additional Comments | |
| Year of Identification | 2005 |
| Year of Validation | 2007 |


