| ProGlyProt ID | BC144 |
| Organism Information |
| Organism Name | Flavobacterium meningosepticum (Elizabethkingia meningoseptica) |
| Domain | Bacteria |
| Classification | Family: Flavobacteriaceae
Order: "Flavobacteriales"
Class: Flavobacteria
Division or phylum: "Bacteroidetes" |
| Taxonomic ID (NCBI) | 238 |
| Genome Sequence (s) |
| EMBL | L06332 |
| Gene Information |
| Gene Name | endOF3 |
| NCBI Gene ID | |
| Protein Information |
| Protein Name | Endo-β-N-acetylglucosaminidase F3 |
| UniProtKB/SwissProt ID | P36913 |
| NCBI RefSeq | |
| EMBL-CDS | AAA24924.1 |
| UniProtKB Sequence | >sp|P36913|EBA3_FLAME Endo-beta-N-acetylglucosaminidase F3 OS=Flavobacterium meningosepticum GN=endOF3 PE=1 SV=1
MKKIFFAQCSILLLMLGSCSKMTEDMTPESVNKEASVKSATALAGSNGVCIAYYITDGRN
PTFKLKDIPDKVDMVILFGLKYWSLQDTTKLPGGTGMMGSFKSYKDLDTQIRSLQSRGIK
VLQNIDDDVSWQSSKPGGFASAAAYGDAIKSIVIDKWKLDGISLDIEHSGAKPNPIPTFP
GYAATGYNGWYSGSMAATPAFLNVISELTKYFGTTAPNNKQLQIASGIDVYAWNKIMENF
RNNFNYIQLQSYGANVSRTQLMMNYATGTNKIPASKMVFGAYAEGGTNQANDVEVAKWTP
TQGAKGGMMIYTYNSNVSYANAVRDAVKN
|
| Sequence length | 329 AA |
| Subcellular Location | Periplasm (secreted) |
| Function | Endohydrolysis of the di-N-acetylchitobiosyl unit in high-mannose glycopeptides and glycoproteins. Hydrolyzes bi- and triantennary glycans. The presence of a core-bound fucose greatly augments endo F3 activity on biantennary and, presumably, triantennary oligosaccharides. EC= 3.2.1.96. |
| Protein Structure |
| PDB ID | 1EOK, 1EOM |
| Glycosylation Status |
| Glycosylation Type | O (Thr) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | (Signal peptide: 1-39) T88
|
| Experimentally Validated Glycosite(s ) in Mature Protein | T49
|
| Glycosite(s) Annotated Protein Sequence | >sp|P36913|EBA3_FLAME Endo-beta-N-acetylglucosaminidase F3 OS=Flavobacterium meningosepticum GN=endOF3 PE=1 SV=1
MKKIFFAQCSILLLMLGSCSKMTEDMTPESVNKEASVKSATALAGSNGVCIAYYITDGRN
PTFKLKDIPDKVDMVILFGLKYWSLQDT*(88)TKLPGGTGMMGSFKSYKDLDTQIRSLQSRGIK
VLQNIDDDVSWQSSKPGGFASAAAYGDAIKSIVIDKWKLDGISLDIEHSGAKPNPIPTFP
GYAATGYNGWYSGSMAATPAFLNVISELTKYFGTTAPNNKQLQIASGIDVYAWNKIMENF
RNNFNYIQLQSYGANVSRTQLMMNYATGTNKIPASKMVFGAYAEGGTNQANDVEVAKWTP
TQGAKGGMMIYTYNSNVSYANAVRDAVKN
|
| Sequence Around Glycosites (21 AA) | FGLKYWSLQDTTKLPGGTGMM |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 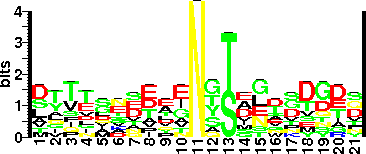 |
| Technique(s) used for Glycosylation Detection | Mass shift detected on SDS-polyacrylamide gel and TFA (trifluoroacetic acid) hydrolysis of intact glycopeptide followed by HPAEC chromatography on a PA-1 column |
| Technique(s) used for Glycosylated Residue(s) Detection | Edman degradation and collision activated dissociation (CAD) mass spectrometry |
| Protein Glycosylation- Implication | Glycosylation may contribute towards protein stability |
| Glycan Information |
| Glycan Annotation | Linkage: Man- Thr.
Branched, acidic, heptasaccharide (1244 Da) containing 3 different uronyl analogs, a methylated Rham and Man, a Glc, and a reducing terminal Man. Only pyranose ring forms were detected [(2-OMe)Man1-4GlcNAcU1-4GlcU1-4Glc1-4(2-OMe)GlcU-4[(2-OMe)Rham1-2]Man]. |
| Technique(s) used for Glycan Identification | ESI-MS (electrospray ionization mass spectrometry), CID (collision-induced dissociation), and a combination of isotopic labeling, composition and methylation analysis. |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | |
| OST UniProtKB/ SwissProt ID | |
| OST NCBI RefSeq | |
| OST EMBL-CDS | |
| OST UniProtKB Sequence | |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Waddling, C.A., Plummer, T.H., Jr., Tarentino, A.L. and Van Roey, P. (2000) Structural basis for the substrate specificity of endo-beta-N-acetylglucosaminidase F(3). Biochemistry, 39, 7878-7885. [PubMed: 10891067]
2) Plummer, T.H., Jr., Tarentino, A.L. and Hauer, C.R. (1995) Novel, specific O-glycosylation of secreted Flavobacterium meningosepticum proteins. Asp-Ser and Asp-Thr-Thr consensus sites. J Biol Chem, 270, 13192-13196. [PubMed: 7768916]
3) Reinhold, B.B., Hauer, C.R., Plum |
| Additional Comments | Identified glycosylation Sequon features: Consensus sequon observed Asp-Ser (DS) or Asp-Thr-Thr (DTT), at turns.
DT alone may not serve as a sequon. Interestingly, the experimentally examined other five nonglycosylated DT sequons were followed by N, D and Q.
Post translational modification of 1.2-kDa was detected by MS in 1993 (Ref. no. 4). The protein migrated slowly on SDS-PAGE (compared to its theoretical weight) and was found to be heterogeneous by MS. |
| Year of Identification | 1995 |
| Year of Validation | 1995 |


