| ProGlyProt ID | BC138 |
| Organism Information |
| Organism Name | Desulfovibrio gigas |
| Domain | Bacteria |
| Classification | Family: Desulfovibrionaceae
Order: Desulfovibrionales
Class: Deltaproteobacteria or Deltabacteria
Division or phylum: "Proteobacteria" |
| Taxonomic ID (NCBI) | 879 |
| Genome Sequence (s) |
| EMBL | |
| Gene Information |
| Gene Name | hcgA |
| NCBI Gene ID | |
| Protein Information |
| Protein Name | HmcA |
| UniProtKB/SwissProt ID | |
| NCBI RefSeq | |
| EMBL-CDS | |
| UniProtKB Sequence | >1Z1N:X|PDBID|CHAIN|SEQUENCE
MTQRKRAARWVGIPCAILFLTVPFISATASTPGPASTAEPKVDAIVIDTAAVFGKLEQPGVVFYHEKHTTALEKMAKDCT
SCHVETEGKLSFKFARTVDPTSKNAMAEQYHANCMACHEKVVGSYPTAPQAAECKRCHVGPGVEGATVTPKPSLDLNLHG
RHVVAEAKRLQVKEDESCKACHHTYDEAQKKLVYAKGEEGSCVYCHKQEPLPSPVQQDRVVPSTRDASHESCVNCHLSTR
KAQTESGPVLCVGCHTAEAQAAWKKTAETPRLFRGQPDATLLVAGAATANGTVDVNWAAAGPGPVAFDHKAHEGFVGNCV
TCHHPTQTGGSLAACGVACHTTTGSKDGNFVTTAQSAHQLGVTTSCVGCHTTQANARKECAGCHAPMQKTALSQNSCIQC
HEAGFPTSGTQTLGKEEREATAAKILAAKDEKPKTVPLENVPEKLTLNYMDEKGDEWQAAEFPHRKIYQKLVEEAAKSPM
ANHFHGDALTMCSGCHHNAKPSLNPPKCASCHSKPFQERTANQPGLKGAFHNQCIGCHQEMQVNPKATDCQGCHKPKNSA
|
| Sequence length | 560 AA |
| Subcellular Location | Periplasm |
| Function | A high molecular mass cytochrome that harbours 16 c-type heme groups. Involved in electron transfer from the periplasm to the cytoplasm. The high molecular mass complex Hmc (HmcA–HmcF) from Desulfovibrio sp. has been proposed to be involved in the bridge between periplasmic hydrogen oxidation and cytoplasmic sulphate reduction in Desulfovibrio gigas. |
| Protein Structure |
| PDB ID | 1Z1N |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | N290 (N261 position in the crystal structure corresponds to the N290 in full length protein sequence) |
| Experimentally Validated Glycosite(s ) in Mature Protein | N290 |
| Glycosite(s) Annotated Protein Sequence | >1Z1N:X|PDBID|CHAIN|SEQUENCE
MTQRKRAARWVGIPCAILFLTVPFISATASTPGPASTAEPKVDAIVIDTAAVFGKLEQPGVVFYHEKHTTALEKMAKDCT
SCHVETEGKLSFKFARTVDPTSKNAMAEQYHANCMACHEKVVGSYPTAPQAAECKRCHVGPGVEGATVTPKPSLDLNLHG
RHVVAEAKRLQVKEDESCKACHHTYDEAQKKLVYAKGEEGSCVYCHKQEPLPSPVQQDRVVPSTRDASHESCVNCHLSTR
KAQTESGPVLCVGCHTAEAQAAWKKTAETPRLFRGQPDATLLVAGAATAN*(290)GTVDVNWAAAGPGPVAFDHKAHEGFVGNCV
TCHHPTQTGGSLAACGVACHTTTGSKDGNFVTTAQSAHQLGVTTSCVGCHTTQANARKECAGCHAPMQKTALSQNSCIQC
HEAGFPTSGTQTLGKEEREATAAKILAAKDEKPKTVPLENVPEKLTLNYMDEKGDEWQAAEFPHRKIYQKLVEEAAKSPM
ANHFHGDALTMCSGCHHNAKPSLNPPKCASCHSKPFQERTANQPGLKGAFHNQCIGCHQEMQVNPKATDCQGCHKPKNSA
|
| Sequence Around Glycosites (21 AA) | TLLVAGAATANGTVDVNWAAA |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 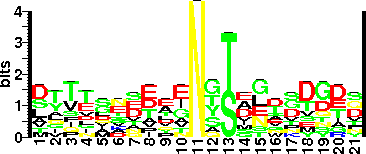 |
| Technique(s) used for Glycosylation Detection | Crystallographic analysis (electron density maps), GlycoProfile III carbohydrate detection kit (using periodic acid as oxidizing agent), MALDI-TOF(matrix-assisted laser desorption/ionization time-of-flight) |
| Technique(s) used for Glycosylated Residue(s) Detection | Crystallographic analysis (electron density maps) |
| Protein Glycosylation- Implication | Carbohydrate could be acting as an anchor of the protein to the phospholipidic membrane. The carbohydrate bound to HmcA may also contribute to the maintenance of the protein conformation and stability as well as to a protection mechanism against proteases. |
| Glycan Information |
| Glycan Annotation | Trisaccharide (NAG,NAA,any epimer of NAG); AllNacGlcNAc-Asn linkage; NAA is (epimer of NAG) N-acetylallosamine. |
| Technique(s) used for Glycan Identification | Crystallographic analysis (electron density maps) |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | |
| OST UniProtKB/ SwissProt ID | |
| OST NCBI RefSeq | |
| OST EMBL-CDS | |
| OST UniProtKB Sequence | |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Santos-Silva, T., Dias, J.M., Dolla, A., Durand, M.C., Goncalves, L.L., Lampreia, J., Moura, I. and Romao, M.J. (2007) Crystal structure of the 16 heme cytochrome from Desulfovibrio gigas: a glycosylated protein in a sulphate-reducing bacterium. J Mol Biol, 370, 659-673. [PubMed: 17531266] |
| Additional Comments | |
| Year of Identification | 2007 |
| Year of Validation | 2007 |


