| ProGlyProt ID | BC137 |
| Organism Information |
| Organism Name | Clostridium difficile strain 630 |
| Domain | Bacteria |
| Classification | Family: Clostridiaceae
Order: Clostridiales
Class: Clostridia
Division or phylum: "Firmicutes" |
| Taxonomic ID (NCBI) | 272563 |
| Genome Sequence (s) |
| GeneBank | AM180355.1 |
| EMBL | AM180355 |
| Gene Information |
| Gene Name | fliC (CD0239) |
| NCBI Gene ID | 4916757 |
| GenBank Gene Sequence | 4916757 |
| Protein Information |
| Protein Name | FliC (Flagellin subunit) |
| UniProtKB/SwissProt ID | Q18CX7 |
| NCBI RefSeq | YP_001086707.1 |
| EMBL-CDS | CAJ67060.1 |
| UniProtKB Sequence | >tr|Q18CX7|Q18CX7_CLOD6 Flagellin subunit OS=Clostridium difficile (strain 630) GN=fliC PE=4 SV=1
MRVNTNVSALIANNQMGRNVNGQSKSMEKLSSGVRIKRAADDAAGLAISEKMRAQIKGLD
QAGRNVQDGISVVQTAEGSLEETGNILQRMRTLSLQSANEINNTEEREKIADELTQLKDE
IERISSSTEFNGKKLLDGTSSTIRLQVGASYGTNVSGTSNNNNEIKIQLVNTASIMASAG
ITTASIGSMKAGGTTGTDAAKTMVSSLDAALKSLNSSRAKLGAQQNRLESTQNNLNNTLE
NVTAAESRIRDTDVASEMVNLSKMNILVQASQSMLAQANQQPQGVLQLLG
|
| Sequence length | 290 AA |
| Subcellular Location | Surface associated |
| Function | Forms the filaments of bacterial flagella. |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | O (Ser/Thr) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | S141, S174, T183, S188, S205 |
| Experimentally Validated Glycosite(s ) in Mature Protein | S141, S174, T183, S188, S205 |
| Glycosite(s) Annotated Protein Sequence | >tr|Q18CX7|Q18CX7_CLOD6 Flagellin subunit OS=Clostridium difficile (strain 630) GN=fliC PE=4 SV=1
MRVNTNVSALIANNQMGRNVNGQSKSMEKLSSGVRIKRAADDAAGLAISEKMRAQIKGLD
QAGRNVQDGISVVQTAEGSLEETGNILQRMRTLSLQSANEINNTEEREKIADELTQLKDE
IERISSSTEFNGKKLLDGTSS*(141)TIRLQVGASYGTNVSGTSNNNNEIKIQLVNTAS*(174) IMASAG
ITT*(183)ASIGS*(188)MKAGGTTGTDAAKTMVS*(205)SLDAALKSLNSSRAKLGAQQNRLESTQNNLNNTLE
NVTAAESRIRDTDVASEMVNLSKMNILVQASQSMLAQANQQPQGVLQLLG
|
| Sequence Around Glycosites (21 AA) | NGKKLLDGTSSTIRLQVGASY
EIKIQLVNTASIMASAGITTA
ASIMASAGITTASIGSMKAGG
SAGITTASIGSMKAGGTTGTD
TGTDAAKTMVSSLDAALKSLN
|
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 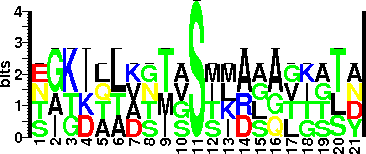 |
| Technique(s) used for Glycosylation Detection | Intact mass analysis with QTOF2-MS (hybrid quadrupole time of flight mass spectrometry) |
| Technique(s) used for Glycosylated Residue(s) Detection | Electron transfer dissociation (ETD) MS |
| Protein Glycosylation- Implication | Glycosylation of the flagellin protein is required for proper assembly and consequent motility. |
| Glycan Information |
| Glycan Annotation | (398-Da glycan) O-linked HexNAc residue, to which a methylated aspartic acid is linked via a phosphate bond. Flagellins from a number of C. difficile isolates from more recent outbreaks are modified in O linkage with a heterogeneous glycan containing up to five monosaccharide residues with masses of 204 (HexNAc), 146 (deoxyhexose), 160 (methylated deoxyhexose), and 192 (heptose). |
| Technique(s) used for Glycan Identification | MS/MS (tandem mass spectrometry), nESI-feCID-MS/MS (nano-electrospray ionization–front-end collision-induced dissociation MS/MS) analyses |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | |
| OST UniProtKB/ SwissProt ID | |
| OST NCBI RefSeq | |
| OST EMBL-CDS | |
| OST UniProtKB Sequence | |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | CD0240 glycosyltransferase. Its insertional mutagenesis abolished the flagellar filament production at the cell surface. |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Twine, S.M., Reid, C.W., Aubry, A., McMullin, D.R., Fulton, K.M., Austin, J. and Logan, S.M. (2009) Motility and flagellar glycosylation in Clostridium difficile. J Bacteriol, 191, 7050-7062. [PubMed: 19749038] |
| Additional Comments | |
| Year of Identification | 2009 |
| Year of Validation | 2009 |


