| ProGlyProt ID | BC128 | | Organism Information | | Organism Name | Campylobacter jejuni subsp. jejuni 81-176 | | Domain | Bacteria | | Classification | Family: Campylobacteraceae
Order: Campylobacterales
Class: Epsilonproteobacteria
Division or phylum: "Proteobacteria" | | Taxonomic ID (NCBI) | 354242 | | Genome Sequence (s) | | GeneBank | CP000538.1 | | EMBL | CP000538 | | Gene Information | | Gene Name | flaA (CJJ81176_1339) | | NCBI Gene ID | 4682159 | | GenBank Gene Sequence | 4682159 | | Protein Information | | Protein Name | Flagellin (FlaA) | | UniProtKB/SwissProt ID | Q2M5R2 | | NCBI RefSeq | YP_001000997.1 | | EMBL-CDS | EAQ72691.1 | | UniProtKB Sequence | >tr|Q2M5R2|Q2M5R2_CAMJJ FlaA OS=Campylobacter jejuni subsp. jejuni serotype O:23/36 (strain 81-176) GN=flaA PE=4 SV=1
MGFRINTNVAALNAKANSDLNAKSLDASLSRLSSGLRINSAADDASGMAIADSLRSQANT
LGQAISNGNDALGILQTADKAMDEQLKILDTIKTKATQAAQDGQSLKTRTMLQADINKLM
EELDNIANTTSFNGKQLLSGNFTNQEFQIGASSNQTVKATIGATQSSKIGVTRFETGAQS
FTSGVVGLTIKNYNGIEDFKFDNVVISTSVGTGLGALAEEINKSADKTGVRATYDVKTTG
VYAIKEGTTSQEFAINGVTIGKIEYKDGDGNGSLISAINAVKDTTGVQASKDENGKLVLT
SADGRGIKITGDIGVGSGILANQKENYGRLSLVKNDGRDINISGTNLSAIGMGTTDMISQ
SSVSLRESKGQISATNADAMGFNSYKGGGKFVFTQNVSSISAFMSAQGSGFSRGSGFSVG
SGKNLSVGLSQGIQIISSAASMSNTYVVSAGSGFSSGSGNSQFAALKTTAANTTDETAGV
TTLKGAMAVMDIAETAITNLDQIRADIGSIQNQVTSTINNITVTQVNVKAAESQIRDVDF
ASESANYSKANILAQSGSYAMAQANSSQQNVLRLLQ
| | Sequence length | 576 AA | | Subcellular Location | Secreted | | Function | It is the subunit protein which is polymerized into the flagellar filaments. Motility mediated by flagella is essential for virulence. | | Protein Structure | | PDB ID | | | Glycosylation Status | | Glycosylation Type | O (Ser) linked | | Experimentally Validated Glycosite(s) in Full Length Protein | S207, S343, S348, T394, S398, S401, S405, S409, S418, S426, S430, S437, S441, S449, S452, S455, S458, S461, T482 | | Experimentally Validated Glycosite(s ) in Mature Protein | S206, S342, S347, T393, S397, S400, S404, S408, S417, S425, S429, S436, S440, S448, S451, S454, S457, S460, T481 | | Glycosite(s) Annotated Protein Sequence | >tr|Q2M5R2|Q2M5R2_CAMJJ FlaA OS=Campylobacter jejuni subsp. jejuni serotype O:23/36 (strain 81-176) GN=flaA PE=4 SV=1
MGFRINTNVAALNAKANSDLNAKSLDASLSRLSSGLRINSAADDASGMAIADSLRSQANT
LGQAISNGNDALGILQTADKAMDEQLKILDTIKTKATQAAQDGQSLKTRTMLQADINKLM
EELDNIANTTSFNGKQLLSGNFTNQEFQIGASSNQTVKATIGATQSSKIGVTRFETGAQS
FTSGVVGLTIKNYNGIEDFKFDNVVIS*(207)TSVGTGLGALAEEINKSADKTGVRATYDVKTTG
VYAIKEGTTSQEFAINGVTIGKIEYKDGDGNGSLISAINAVKDTTGVQASKDENGKLVLT
SADGRGIKITGDIGVGSGILANQKENYGRLSLVKNDGRDINIS*(343)GTNLS*(348)AIGMGTTDMISQ
SSVSLRESKGQISATNADAMGFNSYKGGGKFVFT*(394)QNVS*(398)SIS*(401)AFMS*(405)AQGS*(409)GFSRGSGFS*(418)VGSGKNLS*(426)VGLS*(430) QGIQIIS*(437)SAAS*(441) MSNTYVVS*(449)AGS*(452) GFS*(455)SGS*(458) GNS*(461)QFAALKTTAANTTDETAGVTT*(482)LKGAMAVMDIAETAITNLDQIRADIGSIQNQVTSTINNITVTQVNVKAAESQIRDVDFASESANYSKANILAQSGSYAMAQANSSQQNVLRLLQ
| | Sequence Around Glycosites (21 AA) | EDFKFDNVVISTSVGTGLGAL
VKNDGRDINISGTNLSAIGMG
RDINISGTNLSAIGMGTTDMI
SYKGGGKFVFTQNVSSISAFM
GGKFVFTQNVSSISAFMSAQG
FVFTQNVSSISAFMSAQGSGF
QNVSSISAFMSAQGSGFSRGS
SISAFMSAQGSGFSRGSGFSV
GSGFSRGSGFSVGSGKNLSVG
GFSVGSGKNLSVGLSQGIQII
GSGKNLSVGLSQGIQIISSAA
VGLSQGIQIISSAASMSNTYV
QGIQIISSAASMSNTYVVSAG
AASMSNTYVVSAGSGFSSGSG
MSNTYVVSAGSGFSSGSGNSQ
TYVVSAGSGFSSGSGNSQFAA
VSAGSGFSSGSGNSQFAALKT
GSGFSSGSGNSQFAALKTTAA
| | Glycosite Sequence Logo | seqlogo | | Glycosite Sequence Logo | 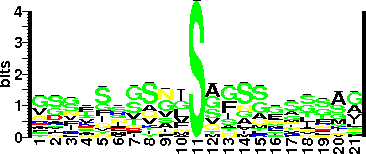 | | Technique(s) used for Glycosylation Detection | ESMS (electrospray mass spectrometry); 10%- 6.5 kDa- mass excess detected | | Technique(s) used for Glycosylated Residue(s) Detection | A combination of μLC-ESMS (liquid chromatography-electrospray mass spectrometry) and MS-MS (tandem mass spectrometry) analyses; 10 out of 19 glycosylation sites were defined using nano-ESMS after base (NH4OH)-catalyzed β-elimination. | | Protein Glycosylation- Implication | Glycosylation is required for flagellar filament formation. Certain glycans mediate filament-filament interactions resulting in AAG (autoagglutination) and other glycans appear to be critical for structural subunit-subunit interactions within the filament. Modification with pseudaminic acid and derivatives is essential for targeting and/or secretion of flagellin. Also, specific structural modifications to the flagellin glycoform have been shown to be involved in the biological fitness of C. jeju | | Glycan Information | | Glycan Annotation | Glycan represents 10% of the total mass of the protein.
Major glycan is pseudaminic acid and its derivatives, Pse5Pr7Pr, Pse5Ac7Ac8OAc, Pse5Am7Ac.
Pse5Ac7Ac (5,7-diacetamido-3,5,7,9 - tetradeoxy-L-glycero-L-manno- nonulosonic acid), with 5-acetamidino pseudaminic acid (Pse5Am7Ac) and 5,7-N-(2,3-dihydroxyproprionyl)-pseudaminic acid (Pse5Pr7Pr) are also present. In addition, novel glycans, Pse5Am7Ac8GlnAc and Pse5Ac7Ac8OAc, have also been found. S398 and S405 carry Pse5Pr7Pr moiety whil | | Technique(s) used for Glycan Identification | Nano-ESMS and NMR analysis of HPLC fractions of trypsin digested glycopeptides including COSY(correlated spectroscopy) and NOESY (nuclear Overhauser effect spectroscopy). | | Protein Glycosylation linked (PGL) gene(s) | | OST Gene Name | | | OST NCBI Gene ID | | | OST GenBank Gene Sequence | | | OST Protein Name | | | OST UniProtKB/ SwissProt ID | A1W0B2 | | OST NCBI RefSeq | | | OST EMBL-CDS | | | OST UniProtKB Sequence | | | OST EC Number (BRENDA) | | | OST Genome Context | | | Characterized Accessory Gene(s) | PseA, PseB, PseC, PseF, PseG, PseH, PseI are the enzymes required for CMP-Pse5NAc7NAc/CMP-Pse5NAc7Am production. PseB and PseC (dehydratase/aminotransferase) catalyze the first two steps of Pse5NAc7NAc (Pse) synthesis. PseI is the Pse synthase. | | PGL Additional Links | CAZy | | Literatures | | Reference(s) | 1) Maita, N., Nyirenda, J., Igura, M., Kamishikiryo, J. and Kohda, D. (2010) Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J Biol Chem, 285, 4941-4950. [PubMed: 20007322]
2) Ewing, C.P., Andreishcheva, E. and Guerry, P. (2009) Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J Bacteriol, 191, 7086-7093. [PubMed: 19749047]
3) McNally, D.J., Hui, J.P., Aubry, A.J., Mui, K.K., Guerry, P., Brisson, J.R., Logan, | | Additional Comments | Pseudaminic acid (Pse5Ac7Ac) has also been identified in the LPS of bacteria.
Pseudomonas pilin also carries pseudaminic acid like glycan (ProGlycProt ID BC166).
Sequon features: No specific sequence features except that all but one of total 19 O-glycosylated residues are restricted to the hydrophobic central, surface-exposed domain of flagellin when folded in the filament. It has been suggested that the local hydrophobicity upstream of Ser/Thr residues partially influences the site | | Year of Identification | 2001 | | Year of Validation | 2001 |
|


