| ProGlyProt ID | BC125 |
| Organism Information |
| Organism Name | Campylobacter jejuni NCTC 11168 serotype O:2 |
| Domain | Bacteria |
| Classification | Family: Campylobacteraceae
Order: Campylobacterales
Class: Epsilonproteobacteria
Division or phylum: "Proteobacteria" |
| Taxonomic ID (NCBI) | 197 |
| Genome Sequence (s) |
| GeneBank | AL111168.1 |
| EMBL | AL111168 |
| Gene Information |
| Gene Name | Cj0289c |
| NCBI Gene ID | 904613 |
| GenBank Gene Sequence | 904613 |
| Protein Information |
| Protein Name | PEB3 |
| UniProtKB/SwissProt ID | Q0PBL7 |
| NCBI RefSeq | YP_002343730.1 |
| EMBL-CDS | CAL34442.1 |
| UniProtKB Sequence | >tr|Q0PBL7|Q0PBL7_CAMJE Major antigenic peptide PEB3 OS=Campylobacter jejuni GN=peb3 PE=1 SV=1
MKKIITLFGACALAFSMANADVNLYGPGGPHTALKDIANKYSEKTGVKVNVNFGPQATWF
EKAKKDADILFGASDQSALAIASDFGKDFNVSKIKPLYFREAIILTQKGNPLKIKGLKDL
ANKKVRIVVPEGAGKSNTSGTGVWEDMIGRTQDIKTIQNFRNNIVAFVPNSGSARKLFAQ
DQADAWITWIDWSKSNPDIGTAVAIEKDLVVYRTFNVIAKEGASKETQDFIAYLSSKEAK
EIFKKYGWRE
|
| Sequence length | 250 AA |
| Subcellular Location | Surface |
| Function | Functions as an adhesin ; also a transport protein that may function in utilization of 3-phosphoglycerate or other phosphate-containing molecules from the host. |
| Protein Structure |
| PDB ID | 2HXW |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | N90 |
| Experimentally Validated Glycosite(s ) in Mature Protein | N90 |
| Glycosite(s) Annotated Protein Sequence | >tr|Q0PBL7|Q0PBL7_CAMJE Major antigenic peptide PEB3 OS=Campylobacter jejuni GN=peb3 PE=1 SV=1
MKKIITLFGACALAFSMANADVNLYGPGGPHTALKDIANKYSEKTGVKVNVNFGPQATWF
EKAKKDADILFGASDQSALAIASDFGKDFN*(90)VSKIKPLYFREAIILTQKGNPLKIKGLKDL
ANKKVRIVVPEGAGKSNTSGTGVWEDMIGRTQDIKTIQNFRNNIVAFVPNSGSARKLFAQ
DQADAWITWIDWSKSNPDIGTAVAIEKDLVVYRTFNVIAKEGASKETQDFIAYLSSKEAK
EIFKKYGWRE
|
| Sequence Around Glycosites (21 AA) | AIASDFGKDFNVSKIKPLYFR |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 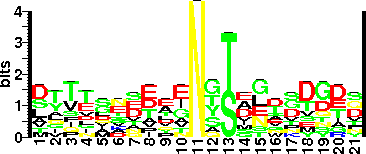 |
| Technique(s) used for Glycosylation Detection | SBA (soybean agglutinin) lectin-agarose affinity chromatography |
| Technique(s) used for Glycosylated Residue(s) Detection | CapLC-MS/MS (capillary liquid chromatography-tandem mass spectrometry) |
| Protein Glycosylation- Implication | |
| Glycan Information |
| Glycan Annotation | Linkage: Bac-Asn.
1406 Da heptasaccharide composed of GalNAc-α1,4-GalNAc-α1,4-[Glcβ1,3-]GalNAc-α1,4-GalNAc-α1,4-GalNAc-α1,3-Bac-β1,N-Asn-Xaa, where Bac is bacillosamine, 2,4-diacetamido-2,4,6-trideoxyglucopyranose. |
| Technique(s) used for Glycan Identification | 1H, 13C NMR spectroscopy- one dimensional TOCSY (total correlation spectroscopy) and NOESY (nuclear Overhauser effect spectroscopy); HMBC (heteronuclear multiple bond coherence), HMQC (heteronuclear multiple quantum correlation) spectra. |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | pglB (Cj1126c) |
| OST NCBI Gene ID | 905417 |
| OST GenBank Gene Sequence | 905417 |
| OST Protein Name | PglB |
| OST UniProtKB/ SwissProt ID | Q0P9C8 |
| OST NCBI RefSeq | YP_002344519.1 |
| OST EMBL-CDS | CAL35243.1 |
| OST UniProtKB Sequence | >tr|Q0P9C8|Q0P9C8_CAMJE Oligosaccharide transferase to N-glycosylate proteins OS=Campylobacter jejuni GN=pglB PE=4 SV=1
MLKKEYLKNPYLVLFAMIVLAYVFSVFCRFYWVWWASEFNEYFFNNQLMIISNDGYAFAE
GARDMIAGFHQPNDLSYYGSSLSTLTYWLYKITPFSFESIILYMSTFLSSLVVIPIILLA
NEYKRPLMGFVAALLASVANSYYNRTMSGYYDTDMLVIVLPMFILFFMVRMILKKDFFSL
IALPLFIGIYLWWYPSSYTLNVALIGLFLIYTLIFHRKEKIFYIAVILSSLTLSNIAWFY
QSAIIVILFALFALEQKRLNFMIIGILGSATLIFLILSGGVDPILYQLKFYIFRSDESAN
LTQGFMYFNVNQTIQEVENVDFSEFMRRISGSEIVFLFSLFGFVWLLRKHKSMIMALPIL
VLGFLALKGGLRFTIYSVPVMALGFGFLLSEFKAILVKKYSQLTSNVCIVFATILTLAPV
FIHIYNYKAPTVFSQNEASLLNQLKNIANREDYVVTWWDYGYPVRYYSDVKTLVDGGKHL
GKDNFFPSFSLSKDEQAAANMARLSVEYTEKSFYAPQNDILKSDILQAMMKDYNQSNVDL
FLASLSKPDFKIDTPKTRDIYLYMPARMSLIFSTVASFSFINLDTGVLDKPFTFSTAYPL
DVKNGEIYLSNGVVLSDDFRSFKIGDNVVSVNSIVEINSIKQGEYKITPIDDKAQFYIFY
LKDSAIPYAQFILMDKTMFNSAYVQMFFLGNYDKNLFDLVINSRDAKVFKLKI |
| OST EC Number (BRENDA) | 2.4.1.119 |
| OST Genome Context | Q0P9C8 |
| Characterized Accessory Gene(s) | PglA, PglJ, PglH, PglI, PglC are glycosyltransferases involved in the heptasaccharide assembly. PglFED are the bacillosamine biosynthetic enzymes. PglK is a flippase. |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Maita, N., Nyirenda, J., Igura, M., Kamishikiryo, J. and Kohda, D. (2010) Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J Biol Chem, 285, 4941-4950. [PubMed: 20007322]
2) Rangarajan, E.S., Bhatia, S., Watson, D.C., Munger, C., Cygler, M., Matte, A. and Young, N.M. (2007) Structural context for protein N-glycosylation in bacteria: The structure of PEB3, an adhesin from Campylobacter jejuni. Protein Sci, 16, 990-995. [PubMed: 17456748]
3) Olivi |
| Additional Comments | Sequon feature: The consensus sequence for the C jejuni PglB is D/E-X'-N-X"-S/T (where X' and X" stand for any amino acid except proline) which is glycosylated when located in flexible, exposed structural elements. Tripeptide sequence NLT too has been found to be modified in vitro. |
| Year of Identification | 2002 |
| Year of Validation | 2002 |


