| ProGlyProt ID | BC124 |
| Organism Information |
| Organism Name | Campylobacter jejuni NCTC 11168 serotype O:2 |
| Domain | Bacteria |
| Classification | Family: Campylobacteraceae
Order: Campylobacterales
Class: Epsilonproteobacteria
Division or phylum: "Proteobacteria" |
| Taxonomic ID (NCBI) | 197 |
| Genome Sequence (s) |
| GeneBank | AL111168.1 |
| EMBL | AL111168 |
| Gene Information |
| Gene Name | Cj0734c (hisJ) |
| NCBI Gene ID | 905052 |
| GenBank Gene Sequence | 905052 |
| Protein Information |
| Protein Name | HisJ |
| UniProtKB/SwissProt ID | Q46125 |
| NCBI RefSeq | YP_002344152.1 |
| EMBL-CDS | CAL34871.1 |
| UniProtKB Sequence | >sp|Q46125|HISJ_CAMJE Histidine-binding protein OS=Campylobacter jejuni GN=hisJ PE=1 SV=1
MKKFLTAFLVAFTGLFLVACQNTKTENNASNEANTTLTLKVGTAPNYKPFNFKQDSKLTG
FDTDLIEEIAKKNGIEIVWVETNFDGLIPALKSGKIDMIASAMSATDERRQSVDFTKPYY
MSKNLYLKLKNNDSLQTKNDLEGKKIGVQLGTLQENTAKAIKNAQVQSNKDLNIAVLALK
NNKIDAIVADQDTAKGFLAENPELVSFYQETDGGEGFSFAFDKNKQKDIIEIFNKGIDEA
KTDGFYDTLIKKYELE |
| Sequence length | 256 AA |
| Subcellular Location | Periplasm |
| Function | A component of the histidine uptake system. |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | N29 |
| Experimentally Validated Glycosite(s ) in Mature Protein | N29 |
| Glycosite(s) Annotated Protein Sequence | >sp|Q46125|HISJ_CAMJE Histidine-binding protein OS=Campylobacter jejuni GN=hisJ PE=1 SV=1
MKKFLTAFLVAFTGLFLGACSDSKNKESN*(29)ASVELKVGTAPNYKPFNFKQDSKLTG
FDTDLIEEIAKKNGIEIVWVETNFDGLIPALKSGKIDMIASAMSATDERRQSVDFTKPYY
MSKNLYLKLKNNDSLQTKNDLEGKKIGVQLGTLQENTAKAIKNAQVQSNKDLNIAVLALK
NNKIDAIVADQDTAKGFLAENPELVSFYQETDGGEGFSFAFDKNKQKDIIEIFNKGIDEA
KTDGFYDTLIKKYELE
|
| Sequence Around Glycosites (21 AA) | ACSDSKNKESNASVELKVGTA |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 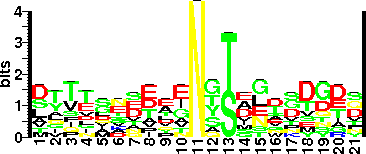 |
| Technique(s) used for Glycosylation Detection | Immunoblot analysis using glycosylation-specific R12 antiserum, SBA (soybean agglutinin) lectin-agarose affinity chromatography, Slow/ aberrant migration on SDS-PAGE |
| Technique(s) used for Glycosylated Residue(s) Detection | Site-directed mutagenesis (N28L) |
| Protein Glycosylation- Implication | |
| Glycan Information |
| Glycan Annotation | |
| Technique(s) used for Glycan Identification | |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | pglB (Cj1126c) |
| OST NCBI Gene ID | 905417 |
| OST GenBank Gene Sequence | 905417 |
| OST Protein Name | PglB |
| OST UniProtKB/ SwissProt ID | Q0P9C8 |
| OST NCBI RefSeq | YP_002344519.1 |
| OST EMBL-CDS | CAL35243.1 |
| OST UniProtKB Sequence | >tr|Q0P9C8|Q0P9C8_CAMJE Oligosaccharide transferase to N-glycosylate proteins OS=Campylobacter jejuni GN=pglB PE=4 SV=1
MLKKEYLKNPYLVLFAMIVLAYVFSVFCRFYWVWWASEFNEYFFNNQLMIISNDGYAFAE
GARDMIAGFHQPNDLSYYGSSLSTLTYWLYKITPFSFESIILYMSTFLSSLVVIPIILLA
NEYKRPLMGFVAALLASVANSYYNRTMSGYYDTDMLVIVLPMFILFFMVRMILKKDFFSL
IALPLFIGIYLWWYPSSYTLNVALIGLFLIYTLIFHRKEKIFYIAVILSSLTLSNIAWFY
QSAIIVILFALFALEQKRLNFMIIGILGSATLIFLILSGGVDPILYQLKFYIFRSDESAN
LTQGFMYFNVNQTIQEVENVDFSEFMRRISGSEIVFLFSLFGFVWLLRKHKSMIMALPIL
VLGFLALKGGLRFTIYSVPVMALGFGFLLSEFKAILVKKYSQLTSNVCIVFATILTLAPV
FIHIYNYKAPTVFSQNEASLLNQLKNIANREDYVVTWWDYGYPVRYYSDVKTLVDGGKHL
GKDNFFPSFSLSKDEQAAANMARLSVEYTEKSFYAPQNDILKSDILQAMMKDYNQSNVDL
FLASLSKPDFKIDTPKTRDIYLYMPARMSLIFSTVASFSFINLDTGVLDKPFTFSTAYPL
DVKNGEIYLSNGVVLSDDFRSFKIGDNVVSVNSIVEINSIKQGEYKITPIDDKAQFYIFY
LKDSAIPYAQFILMDKTMFNSAYVQMFFLGNYDKNLFDLVINSRDAKVFKLKI |
| OST EC Number (BRENDA) | 2.4.1.119 |
| OST Genome Context | Q0P9C8 |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Maita, N., Nyirenda, J., Igura, M., Kamishikiryo, J. and Kohda, D. (2010) Comparative structural biology of eubacterial and archaeal oligosaccharyltransferases. J Biol Chem, 285, 4941-4950. [PubMed: 20007322]
2) Nita-Lazar, M., Wacker, M., Schegg, B., Amber, S. and Aebi, M. (2005) The N-X-S/T consensus sequence is required but not sufficient for bacterial N-linked protein glycosylation. Glycobiology, 15, 361-367. [PubMed: 15574802] |
| Additional Comments | Glycosylation at sequon 28-N-A-S-30 and not at the other potential sequon 132-N-D-S-134 in glycosylation competent recombinant E.coli strain as well as in native host strain suggests for the first time that N-X-S/T sequon is required but not sufficient in case of campylobacter/ bacterial N glycosylation. Seat of glycosylation was found to be periplasm. N127 is experimentally proved (by site directed mutagenesis) as non-glycosylated sequon.
Sequence conflict has been observed at many places. |
| Year of Identification | 2005 |
| Year of Validation | 2005 |


