| ProGlyProt ID | BC110 |
| Organism Information |
| Organism Name | Bacteroides fragilis (strain ATCC 25285 / NCTC 9343) |
| Domain | Bacteria |
| Classification | Family: Bacteroidaceae
Order: "Bacteroidales"
Class: "Bacteroidia"
Division or phylum: "Bacteroidetes" |
| Taxonomic ID (NCBI) | 272559 |
| Genome Sequence (s) |
| GeneBank | CR626927.1 |
| EMBL | CR626927 |
| Gene Information |
| Gene Name | BF0522 |
| NCBI Gene ID | 3286009 |
| GenBank Gene Sequence | 3286009 |
| Protein Information |
| Protein Name | Hypothetical protein |
| UniProtKB/SwissProt ID | Q5LHV3 |
| NCBI RefSeq | YP_210235.1 |
| EMBL-CDS | CAH06277.1 |
| UniProtKB Sequence | >tr|Q5LHV3|Q5LHV3_BACFN Putative uncharacterized protein OS=Bacteroides fragilis (strain ATCC 25285 / NCTC 9343) GN=BF0522 PE=4 SV=1
MKRLIPILLAVFAFAACEKDPDMDKLDNDYLVYTNYDKKADFKQFSTYYIPDSVLVIGDK
KDPEYWKGEAAEAIINAYKENLNSKGFTYTDNKDAADLGIQVSYVQSTYYFTDYGQPEWW
WNYPGYWDAPYWGNWGGWYYPYVVNYSITTNSFLTEIMNLKAPEGEKQKLPVLWSSFLSG
PASYSGKVNQTLVVRAINQSFAQSPYLTNK |
| Sequence length | 210 AA |
| Subcellular Location | Outer membrane (facing surface) |
| Function | It is suggested to be involved in the response to oxidative stress as it was found upregulated 6.1-fold on exposure of B. fragilis to air. |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | O (Ser) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | S53 |
| Experimentally Validated Glycosite(s ) in Mature Protein | S53 |
| Glycosite(s) Annotated Protein Sequence | >tr|Q5LHV3|Q5LHV3_BACFN Putative uncharacterized protein OS=Bacteroides fragilis (strain ATCC 25285 / NCTC 9343) GN=BF0522 PE=4 SV=1
MKRLIPILLAVFAFAACEKDPDMDKLDNDYLVYTNYDKKADFKQFSTYYIPDS*(53)VLVIGDK
KDPEYWKGEAAEAIINAYKENLNSKGFTYTDNKDAADLGIQVSYVQSTYYFTDYGQPEWW
WNYPGYWDAPYWGNWGGWYYPYVVNYSITTNSFLTEIMNLKAPEGEKQKLPVLWSSFLSG
PASYSGKVNQTLVVRAINQSFAQSPYLTNK
|
| Sequence Around Glycosites (21 AA) | KQFSTYYIPDSVLVIGDKKDP |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 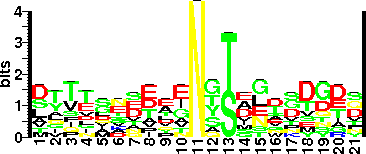 |
| Technique(s) used for Glycosylation Detection | Mass shift detected on SDS-polyacrylamide gel and Western blot (using wild-type and mutant B. fragilis cells for expression, and anti-His tag antibody detection) |
| Technique(s) used for Glycosylated Residue(s) Detection | Site-directed mutagenesis |
| Protein Glycosylation- Implication | Protein glycosylation is central to the physiology of B. fragilis and is necessary for the organism to competitively colonize the mammalian intestine. Deletion of the lfg (protein glycosylation machinery) region results in a substantial growth deficiency in vitro and a complete inability to compete with wild-type bacteria in the mouse intestine. |
| Glycan Information |
| Glycan Annotation | Exogenous fucose. |
| Technique(s) used for Glycan Identification | Lectin (AAL)binding |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | wcfB |
| OST NCBI Gene ID | 3287270 |
| OST GenBank Gene Sequence | 3287270 |
| OST Protein Name | Putative fucosyl transferase |
| OST UniProtKB/ SwissProt ID | Q5LGI6 |
| OST NCBI RefSeq | YP_210702.1 |
| OST EMBL-CDS | CAH06753.1 |
| OST UniProtKB Sequence | >tr|Q5LGI6|Q5LGI6_BACFN Putative fucosyl transferase OS=Bacteroides fragilis (strain ATCC 25285 / NCTC 9343) GN=wcfB PE=4 SV=1
MLYVILRGRLGNNLFQIATAASLTQNFIFCTVNKDQERQVLLYKDSFFKNIKVMKGVPDG
IPYYKEPFHEFSRIPYEEGKDLIIDGYFQSEKYFKRSVVLDLYRITDELRKKIWNICGNI
LEKGETVSIHVRRGDYLKLPHALPFCGKSYYKNAIQYIGEDKIFIICSDDIDWCKKNFIG
KRYYFIENTTPLLDLYIQSLCTHNIISNSSFSWWGAWLNENSNKIVIAPQMWFGISVKLG
VSDLLPVSWVRLPNNYTLGRYCFALYKVVEDYLLNILRLIWKRKKNM |
| OST EC Number (BRENDA) | |
| OST Genome Context | Q5LGI6 |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Fletcher, C.M., Coyne, M.J. and Comstock, L.E. (2011) Theoretical and experimental characterization of the scope of protein O-glycosylation in Bacteroides fragilis. J Biol Chem, 286, 3219-3226. [PubMed: 21115495]
2) Fletcher, C.M., Coyne, M.J., Villa, O.F., Chatzidaki-Livanis, M. and Comstock, L.E. (2009) A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell, 137, 321-331. [PubMed: 19379697] |
| Additional Comments | Glycosylation sequon features: the sequon has an aspartate (D) preceding the glycosylated T or S which is followed by an amino acid with one or more methyl groups (alanine, isoleucine, or leucine; (D)(S/T)(A/I/L/V/M/T). Moreover, none of the 17 unglycosylated S and T residues examined in of BF2494 (excluding two in the signal peptide) have a preceding D, although seven are followed by A, I, or L and one by V. Non methylated amino acids were not tolerated at third position of sequon in BF2494. Il |
| Year of Identification | 2009 |
| Year of Validation | 2011 |


