| ProGlyProt ID | BC106 |
| Organism Information |
| Organism Name | Bacillus subtilis 168 |
| Domain | Bacteria |
| Classification | Family: Bacillaceae
Order: Bacillales
Class: Bacilli (or Firmibacteria)
Division or phylum: "Firmicutes" |
| Taxonomic ID (NCBI) | 1423 |
| Genome Sequence (s) |
| GeneBank | AF014938.1 |
| EMBL | AF014938 |
| Gene Information |
| Gene Name | sunA |
| NCBI Gene ID | 939121 |
| GenBank Gene Sequence | 939121 |
| Protein Information |
| Protein Name | Sublansin |
| UniProtKB/SwissProt ID | P68577 |
| NCBI RefSeq | NP_390031.1 |
| EMBL-CDS | AAC63531.1 |
| UniProtKB Sequence | >sp|P68577|SUNA_BACSU SPBc2 prophage-derived bacteriocin sublancin-168 OS=Bacillus subtilis GN=sunA PE=1 SV=1
MEKLFKEVKLEELENQKGSGLGKAQCAALWLQCASGGTIGCGGGAVACQNYRQFCR |
| Sequence length | 56 AA |
| Subcellular Location | Secreted |
| Function | SPβ prophage-derived bacteriocin sublancin-168. It has antimicrobial activity against Gram-positive bacteria. It is stable at both low and high pH and lacks free thiols. |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | S (Cys) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | (Propeptide: 1-19) C41 |
| Experimentally Validated Glycosite(s ) in Mature Protein | C22 |
| Glycosite(s) Annotated Protein Sequence | >sp|P68577|SUNA_BACSU SPBc2 prophage-derived bacteriocin sublancin-168 OS=Bacillus subtilis GN=sunA PE=1 SV=1
MEKLFKEVKLEELENQKGSGLGKAQCAALWLQCASGGTIGC*(41)GGGAVACQNYRQFCR
|
| Sequence Around Glycosites (21 AA) | LQCASGGTIGCGGGAVACQNY |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 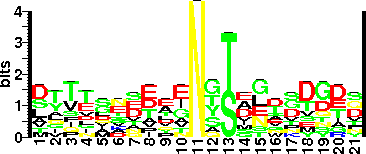 |
| Technique(s) used for Glycosylation Detection | Higher mass observed using mass spectrometry |
| Technique(s) used for Glycosylated Residue(s) Detection | Tandem ESI-MS (electrospray ionization quadrupole-TOF mass spectrometry) analysis after chymotrypsin digestion. |
| Protein Glycosylation- Implication | Glucosylation is essential for its bioactivity. |
| Glycan Information |
| Glycan Annotation | UDP-Glc, UDP-GlcNAc, UDP-Gal, GDP-Man and UDP-Xyl can serve as substrates for SunS GTase but UDP-α-D-glucose is most efficiently used. |
| Technique(s) used for Glycan Identification | GC-MS (gas chromatography-mass spectrometry) analysis after trimethylsilylation |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | |
| OST UniProtKB/ SwissProt ID | |
| OST NCBI RefSeq | |
| OST EMBL-CDS | |
| OST UniProtKB Sequence | |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | SunS is a glycosyltransferase with very relaxed substrate specificity. It shows strong regioselectivity and chemoselectivity for glycosylation of a thiol. |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Oman, T.J., Boettcher, J.M., Wang, H., Okalibe, X.N. and van der Donk, W.A. (2011) Sublancin is not a lantibiotic but an S-linked glycopeptide. Nat Chem Biol, 7, 78-80. [PubMed: 21196935]
2) Stepper, J., Shastri, S., Loo, T.S., Preston, J.C., Novak, P., Man, P., Moore, C.H., Havlicek, V., Patchett, M.L. and Norris, G.E. (2011) Cysteine S-glycosylation, a new post-translational modification found in glycopeptide bacteriocins. FEBS Lett, 585, 645-650. [PubMed: 21251913] |
| Additional Comments | S-linked glycosylation is very rare. It has been observed that cysteine glycosylation leads to the formation of more stable products (both at low and high pH) than does serine glycosylation. |
| Year of Identification | 2011 |
| Year of Validation | 2011 |


