| ProGlyProt ID | BC101 |
| Organism Information |
| Organism Name | Acetogenium kivui (Thermoanaerobacter kivui) |
| Domain | Bacteria |
| Classification | Family: Thermoanaerobacteraceae
Order: Thermoanaerobacterales
Class: Clostridia
Division or phylum: "Firmicutes" |
| Taxonomic ID (NCBI) | 2325 |
| Genome Sequence (s) |
| EMBL | M31069 |
| Gene Information |
| Gene Name | |
| NCBI Gene ID | |
| Protein Information |
| Protein Name | Cell surface protein/ S-layer protein
|
| UniProtKB/SwissProt ID | P22258 |
| NCBI RefSeq | |
| EMBL-CDS | AAA21930.1 |
| UniProtKB Sequence | >sp|P22258|SLAP_ACEKI Cell surface protein OS=Acetogenium kivui PE=1 SV=1
MKNLKKLIAVVSTFALVFSAMAVGFAATTPFTDVKDDAPYASAVARLYALNITNGVGDPK
FGVDQPVTRAQMITFVNRMLGYEDLAEMAKSEKSAFKDVPQNHWAVGQINLAYKLGLAQG
VGNGKFDPNSELRYAQALAFVLRALGFKDLDWPYGYLAKAQDLGLVHGLNLAYNGVIKRG
DLALILDRALEVPMVKYVDGKEVLGEPLISKVATKAEYTVIATNAQDRSVEEGKVAVLDK
DGKLTTINAGLVDFSEYLGKKVIVYSERFGDPVYVAEGDNDVVSFTEGQDSVGTTVYKND
DNKTAIKVDDNAYVLYNGYLTKVSKVTVKEGAEVTIINNNYLIVNGSYDNSTIVYNDVQS
GDKYLNRDSNYELKGTVTVTGAVSKVTDIKANDYIYYGKQYDVNGNVVGTVIYVVRNQVT
GTVTEKSVSGSTYKASIDNVSYTVADNNVWNQLEPGKKVTVILNKDNVIVGISSTTTTTA
VNYAIFKEKSDPFTAWFAKVKLILPDAAEKVFDAVYSDVYDKVNLAEGTIVTYTVDANGK
LNDIQRANDQPFSSASYKADAKVLTEGSTTYYITDNTVLLNNTSDGYKALKLTDLKDATN
LNVKIVADNYNVAKVVVFNNASFVSTTTSTVYAYVTGTADVYVNGSTFTRLTVLENGQTK
TYDANAQLATNYTHKAVVLTLTNAKIANIALPTVASGVKLTNIDQANLRITDTTNKGYLL
DPNFIVVDTNGNLKGLSDITKDTGVNLYTNDVGKVFVIEIVQ
|
| Sequence length | 762 AA |
| Subcellular Location | Surface |
| Function | The S-layer is a paracrystalline mono-layered assembly of proteins which coat the surface of bacteria. This bacterium is covered by a S-layer with hexagonal symmetry.
|
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | O (Tyr) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | Y297, Y516, Y520, Y632 |
| Experimentally Validated Glycosite(s ) in Mature Protein | Y297, Y516, Y520, Y632 |
| Glycosite(s) Annotated Protein Sequence | >sp|P22258|SLAP_ACEKI Cell surface protein OS=Acetogenium kivui PE=1 SV=1
MKNLKKLIAVVSTFALVFSAMAVGFAATTPFTDVKDDAPYASAVARLYALNITNGVGDPK
FGVDQPVTRAQMITFVNRMLGYEDLAEMAKSEKSAFKDVPQNHWAVGQINLAYKLGLAQG
VGNGKFDPNSELRYAQALAFVLRALGFKDLDWPYGYLAKAQDLGLVHGLNLAYNGVIKRG
DLALILDRALEVPMVKYVDGKEVLGEPLISKVATKAEYTVIATNAQDRSVEEGKVAVLDK
DGKLTTINAGLVDFSEYLGKKVIVYSERFGDPVYVAEGDNDVVSFTEGQDSVGTTVY*(297)KND
DNKTAIKVDDNAYVLYNGYLTKVSKVTVKEGAEVTIINNNYLIVNGSYDNSTIVYNDVQS
GDKYLNRDSNYELKGTVTVTGAVSKVTDIKANDYIYYGKQYDVNGNVVGTVIYVVRNQVT
GTVTEKSVSGSTYKASIDNVSYTVADNNVWNQLEPGKKVTVILNKDNVIVGISSTTTTTA
VNYAIFKEKSDPFTAWFAKVKLILPDAAEKVFDAVY*(516)SDVY*(520)DKVNLAEGTIVTYTVDANGK
LNDIQRANDQPFSSASYKADAKVLTEGSTTYYITDNTVLLNNTSDGYKALKLTDLKDATN
LNVKIVADNYNVAKVVVFNNASFVSTTTSTVY*(632)AYVTGTADVYVNGSTFTRLTVLENGQTK
TYDANAQLATNYTHKAVVLTLTNAKIANIALPTVASGVKLTNIDQANLRITDTTNKGYLL
DPNFIVVDTNGNLKGLSDITKDTGVNLYTNDVGKVFVIEIVQ
|
| Sequence Around Glycosites (21 AA) | EGQDSVGTTVYKNDDNKTAIK
DAAEKVFDAVYSDVYDKVNLA
KVFDAVYSDVYDKVNLAEGTI
SFVSTTTSTVYAYVTGTADVY
|
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 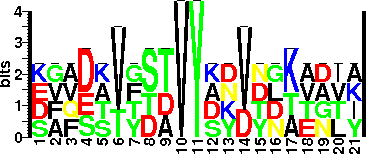 |
| Technique(s) used for Glycosylation Detection | Deglycosylation (trifluoromethanesulfonic acid), aberrant migration on SDS-PAGE. |
| Technique(s) used for Glycosylated Residue(s) Detection | Protease digestion and glycopeptide separation by molecular sieve chromatography and reversed-phase HPLC. Deglycosylated and undeglycosylated peptides were run on 2D PAGE and subjected to N-terminal sequencing post blotting. |
| Protein Glycosylation- Implication | An N terminal SLH sequence of A. kivui S layer glycoprotein is suggested to fold independently and present itself in a modular way that may serve as an anchor to the peptidoglycan. Glycosylated residues are surface exposed and implicated in protease K resistance. |
| Glycan Information |
| Glycan Annotation | Carbohydrate content 8% corresponding to 40 to 50 sugar molecules per protein monomer. Acidic, repetitive heterogenous glycans of varying length containing N-acetylglucosamine, 1,6 linked glucose, 1,4 linked galactosamine and/or unidentified sugar-related component. |
| Technique(s) used for Glycan Identification | Hydrazinolysis followed by amino-acid analysis as well as two-dimensional NMR spectroscopy |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | |
| OST UniProtKB/ SwissProt ID | |
| OST NCBI RefSeq | |
| OST EMBL-CDS | |
| OST UniProtKB Sequence | |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Lupas, A., Engelhardt, H., Peters, J., Santarius, U., Volker, S. and Baumeister, W. (1994) Domain structure of the Acetogenium kivui surface layer revealed by electron crystallography and sequence analysis. J Bacteriol, 176, 1224-1233. [PubMed: 8113161]
2) Peters, J., Rudolf, S., Oschkinat, H., Mengele, R., Sumper, M., Kellermann, J., Lottspeich, F. and Baumeister, W. (1992) Evidence for tyrosine-linked glycosaminoglycan in a bacterial surface protein. Biol Chem Hoppe Seyler, 373, 171-17 |
| Additional Comments | A new recognition sequence for tyrosine linked glycosylation in A. kivui is suggested based on the fact that all glycosylated tyrosine in the sequence are preceded by valine. |
| Year of Identification | 1989 |
| Year of Validation | 1992 |


