| ProGlyProt ID | AC122 |
| Organism Information |
| Organism Name | Sulfolobus acidocaldarius (DSM 639) |
| Domain | Archaea |
| Classification | Family: Sulfolobaceae
Order: Sulfolobales
Class: Thermoprotei or Crenarchaeota
Division or phylum: "Crenarchaeota" |
| Taxonomic ID (NCBI) | 2285 |
| Genome Sequence (s) |
| GeneBank | CP000077.1 |
| EMBL | CP000077 |
| Gene Information |
| Gene Name | thpS (Saci_1714) |
| NCBI Gene ID | 3474479 |
| GenBank Gene Sequence | 3474479 |
| Protein Information |
| Protein Name | Thermopsin |
| UniProtKB/SwissProt ID | P17118 |
| NCBI RefSeq | YP_256313.1 |
| EMBL-CDS | AAY81020.1 |
| UniProtKB Sequence | >sp|P17118|THPS_SULAC Thermopsin OS=Sulfolobus acidocaldarius GN=thpS PE=1 SV=1
MNFKSICLIILLSALIIPYIPQNIYFFPHRNTTGATISSGLYVNPYLYYTSPPAPAGIAS
FGLYNYSGNVTPYVITTNEMLGYVNITSLLAYNREALRYGVDPYSATLQFNIVLSVNTSN
GVYAYWLQDVGQFQTNKNSLTFIDNVWNLTGSLSTLSSSAITGNGQVASAGGGQTFYYDV
GPSYTYSFPLSYIYIINMSYTSNAVYVWIGYEIIQIGQTEYGTVNYYDKITIYQPNIISA
SLMINGNNYTPNGLYYDAELVWGGGGNGAPTSFNSLNCTLGLYYISNGSITPVPSLYTFG
ADTAEAAYNVYTTMNNGVPIAYNGIENLTILTNNFSVILI
|
| Sequence length | 340 AA |
| Subcellular Location | Secreted |
| Function | Thermostable acid protease. It has maximal proteolytic activity at pH 2 and 90 “C.Belongs to the peptidase A5 family. EC= 3.4.23.42. |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | (Signal peptide: 1-28, Propeptide: 28-41) N65, N69 |
| Experimentally Validated Glycosite(s ) in Mature Protein | N24, N28 |
| Glycosite(s) Annotated Protein Sequence | >sp|P17118|THPS_SULAC Thermopsin OS=Sulfolobus acidocaldarius GN=thpS PE=1 SV=1
MNFKSICLIILLSALIIPYIPQNIYFFPHRNTTGATISSGLYVNPYLYYTSPPAPAGIAS
FGLYN*(65)YSGN*(69)VTPYVITTNEMLGYVNITSLLAYNREALRYGVDPYSATLQFNIVLSVNTSN
GVYAYWLQDVGQFQTNKNSLTFIDNVWNLTGSLSTLSSSAITGNGQVASAGGGQTFYYDV
GPSYTYSFPLSYIYIINMSYTSNAVYVWIGYEIIQIGQTEYGTVNYYDKITIYQPNIISA
SLMINGNNYTPNGLYYDAELVWGGGGNGAPTSFNSLNCTLGLYYISNGSITPVPSLYTFG
ADTAEAAYNVYTTMNNGVPIAYNGIENLTILTNNFSVILI
|
| Sequence Around Glycosites (21 AA) | PAGIASFGLYNYSGNVTPYVI
ASFGLYNYSGNVTPYVITTNE
|
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 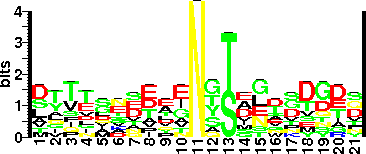 |
| Technique(s) used for Glycosylation Detection | Aberrant SDS-PAGE migration. |
| Technique(s) used for Glycosylated Residue(s) Detection | Edman degradation |
| Protein Glycosylation- Implication | |
| Glycan Information |
| Glycan Annotation | |
| Technique(s) used for Glycan Identification | |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | Saci_1274 |
| OST NCBI Gene ID | 3473075 |
| OST GenBank Gene Sequence | 3473075 |
| OST Protein Name | Conserved membrane protein |
| OST UniProtKB/ SwissProt ID | Q4J9B4 |
| OST NCBI RefSeq | YP_255909.1 |
| OST EMBL-CDS | AAY80616.1 |
| OST UniProtKB Sequence | >tr|Q4J9B4|Q4J9B4_SULAC Conserved membrane protein OS=Sulfolobus acidocaldarius GN=Saci_1274 PE=4 SV=1
MQSTSILARIDKFKFLDAIIIGSLALFSILIRIISITAFPQTINGFDSWYLFYNALLIVK
AGGNWYAVPPDVHAWFPWGYFIELENTIGLPFLVALFSVPFYSIFGQNIVYTLTLVSPIV
LDGIGVVAAFLAVESITNSRVGGYIAAAITAFTPSLTYKNILGSLPKTSWGGVFVLFTIY
FLSLAIKKKKPLYGIPAGIMIFLANITWGGYTYIDISLAIAAFLIVLFNKNDEISAKTLT
ISGITAAFLTSLSPNTIGFMSEVAHGLALLIIPLFLYLDLYLRRVLPKDIVDSKNIVIGA
AIILLVSLVVLGSVAFKVQLIPSRYYAIINPFFQFTVPIDRTVAEYIPQSIAAMIQDFGI
GLFLSIIGIYFLLTRKQDMAGIWLVVLGAASIYGTSEQPYLFNYTIYIVAALAGVAVAEL
FSRFMERKIRIAPILMLTLIGVALLADAGIAVEASYAPQALINSSTSYLTTNYAWISALD
WINQNTPNNAFILSWWDYGYWIHAVGNRTVIDENNTLNGTQIKLMAEMFLNNESFAVNVL
ENDFHLYPYGNPNYTRPVYIVAYDAVTEYIVNNQYPVWFIGYPTNFPGTFIGYTTSLGDI
AKAIGAMTTIAGYNTNSYVNTTYINETASYAAQYNSQLASIIANSLPMAWTPKTYNSLIG
SMFIEAIQSLNQGPVQAPFSISLSQLLQSSSTSLYNPNALLPRVNLMYFKPVYIALFPLS
VTNALGGEAIVYIMVYIYQFVMPNVIIPPTISTA
|
| OST EC Number (BRENDA) | 2.4.1.119 |
| OST Genome Context | Q4J9B4 |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Lin, X. and Tang, J. (1990) Purification, characterization, and gene cloning of thermopsin, a thermostable acid protease from Sulfolobus acidocaldarius. J Biol Chem, 265, 1490-1495. [PubMed: 2104843] |
| Additional Comments | |
| Year of Identification | 1990 |
| Year of Validation | 1990 |


