| ProGlyProt ID | AC121 |
| Organism Information |
| Organism Name | Sulfolobus acidocaldarius (DSM 639) |
| Domain | Archaea |
| Classification | Family: Sulfolobaceae
Order: Sulfolobales
Class: Thermoprotei or Crenarchaeota
Division or phylum: "Crenarchaeota" |
| Taxonomic ID (NCBI) | 2285 |
| Genome Sequence (s) |
| GeneBank | CP000077.1 |
| EMBL | CP000077 |
| Gene Information |
| Gene Name | slaA (Saci_2355) or slp1
|
| NCBI Gene ID | 3472789 |
| GenBank Gene Sequence | 3472789 |
| Protein Information |
| Protein Name | SlaA (S- layer protein) |
| UniProtKB/SwissProt ID | Q4J6E5 |
| NCBI RefSeq | YP_256929.1 |
| EMBL-CDS | AAY81636.1 |
| UniProtKB Sequence | >tr|Q4J6E5|Q4J6E5_SULAC Conserved protein OS=Sulfolobus acidocaldarius GN=Saci_2355 PE=4 SV=1
MNKLVGLLVSSLFLASILIGIAPAITTTALTPPVSAGGIQAYLLTGSGAPASGLVLFVVN
VSNIQVSSSNVTNVISTVVSNIQINAKTENAQTGATTGSVTVRFPTSGYNAYYDSVDKVV
FVVVSFLYPYTTTSVNIPLSYLSKYLPGLLTAQPYDETGAQVTSVSSTPFGSLIDTSTGQ
QILGTNPVLTSYNSYTTQANTNMQEGVVSGTLTSFTLGGQSFSGSTVPVILYAPFIFSNS
PYQAGLYNPMQVNGNLGSLSSEAYYHPVIWGRALINTTLIDTYASGSVPFTFQLNYSVPG
PLTINMAQLAWIASINNLPTSFTYLSYKFSNGYESFLGIISNSTQLTAGALTINPSGNFT
INGKKFYVYLLVVGSTNSTTPVEYVTKLVVEYPSSTNFLPQGVTVTTSSNKYTLPVYEIG
GPAGTTITLTGNWYSTPYTVQITVGSTPTLTNYVSQILLKAVAYEGINVSTTQSPYYSTA
ILSTPPSEISITGSSTITAQGKLTATSASATVNLLTNATLTYENIPLTQYSFNGIIVTPG
YAAINGTTAMAYVIGALYNKTSDYVLSFAGSQEPMQVMNNNLTEVTTLAPFGLTLLAPSV
PATETGTSPLQLEFFTVPSTSYIALVDFGLWGNLTSVTVSAYDTVNNKLSVNLGYFYGIV
IPPSISTAPYNYQNFICPNNYVTVTIYDPDAVLDPYPSGSFTTSSLPLKYGNMNITGAVI
FPGSSVYNPSGVFGYSNFNKGAAVTTFTYTAQSGPFSPVALTGNTNYLSQYADNNPTDNY
YFIQTVNGMPVLMGGLSIVASPVSASLPSSTSSPGFMYLLPSAAQVPSPLPGMATPNYNL
NIYITYKIDGATVGNNMINGLYVASQNTLIYVVPNGSFVGSNIKLTYTTTDYAVLHYFYS
TGQYKVFKTVSVPNVTANLYFPSSTTPLYQLSVPLYLSEPYYGSPLPTYIGLGTNGTSLW
NSPNYVLFGVSAVQQYLGFIKSISVTLSNGTTVVIPLTTSNMQTLFPQLVGQELQACNGT
FQFGISITGLEKLLNLNVQQLNNSILSVTYHDYVTGETLTATTKLVALSTLSLVAKGAGV
VEFLLTAYPYTGNITFAPPWFIAENVVKQPFMTYSDLQFAKTNPSAILSLSTVNITVVGL
GGKASVYYNSTSGQTVITNIYGQTVATLSGNVLPTLTELAAGNGTFTGSLQFTIVPNNTV
VQIPSSLTKTSFAVYTNGSLAIVLNGKAYSLGPAGLFLLPFVTYTGSAIGANATAIITVS
DGVGTSTTQVPITAENFTPIRLAPFQVPAQVPLPNAPKLKYEYNGSIVITPQQQVLKIYV
TSILPYPQEFQIQAFVYEASQFNVHTGSPTAAPVYFSYSAVRAYPALGIGTSVPNLLVYV
QLQGISNLPAGKYVIVLSAVPFAGGPVLSEYPAQLIFTNVTLTQ |
| Sequence length | 1424 AA |
| Subcellular Location | Surface |
| Function | The S-layer has to maintain the cell integrity and stabilize as well as to protect the cell against mechanical and osmotic stresses or extreme pH conditions. It is also predicted that the S-layer has to maintain or even determine the cell shape. |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | (Signal peptide : 1-29) N1042, N1093, N1134, N1197, N1217, N1252, N1276, N1304, N1419
|
| Experimentally Validated Glycosite(s ) in Mature Protein | N1013, N1064, N1105, N1168, N1188, N1223, N1247, N1275, N1390
|
| Glycosite(s) Annotated Protein Sequence | >tr|Q4J6E5|Q4J6E5_SULAC Conserved protein OS=Sulfolobus acidocaldarius GN=Saci_2355 PE=4 SV=1
MNKLVGLLVSSLFLASILIGIAPAITTTALTPPVSAGGIQAYLLTGSGAPASGLVLFVVN
VSNIQVSSSNVTNVISTVVSNIQINAKTENAQTGATTGSVTVRFPTSGYNAYYDSVDKVV
FVVVSFLYPYTTTSVNIPLSYLSKYLPGLLTAQPYDETGAQVTSVSSTPFGSLIDTSTGQ
QILGTNPVLTSYNSYTTQANTNMQEGVVSGTLTSFTLGGQSFSGSTVPVILYAPFIFSNS
PYQAGLYNPMQVNGNLGSLSSEAYYHPVIWGRALINTTLIDTYASGSVPFTFQLNYSVPG
PLTINMAQLAWIASINNLPTSFTYLSYKFSNGYESFLGIISNSTQLTAGALTINPSGNFT
INGKKFYVYLLVVGSTNSTTPVEYVTKLVVEYPSSTNFLPQGVTVTTSSNKYTLPVYEIG
GPAGTTITLTGNWYSTPYTVQITVGSTPTLTNYVSQILLKAVAYEGINVSTTQSPYYSTA
ILSTPPSEISITGSSTITAQGKLTATSASATVNLLTNATLTYENIPLTQYSFNGIIVTPG
YAAINGTTAMAYVIGALYNKTSDYVLSFAGSQEPMQVMNNNLTEVTTLAPFGLTLLAPSV
PATETGTSPLQLEFFTVPSTSYIALVDFGLWGNLTSVTVSAYDTVNNKLSVNLGYFYGIV
IPPSISTAPYNYQNFICPNNYVTVTIYDPDAVLDPYPSGSFTTSSLPLKYGNMNITGAVI
FPGSSVYNPSGVFGYSNFNKGAAVTTFTYTAQSGPFSPVALTGNTNYLSQYADNNPTDNY
YFIQTVNGMPVLMGGLSIVASPVSASLPSSTSSPGFMYLLPSAAQVPSPLPGMATPNYNL
NIYITYKIDGATVGNNMINGLYVASQNTLIYVVPNGSFVGSNIKLTYTTTDYAVLHYFYS
TGQYKVFKTVSVPNVTANLYFPSSTTPLYQLSVPLYLSEPYYGSPLPTYIGLGTNGTSLW
NSPNYVLFGVSAVQQYLGFIKSISVTLSNGTTVVIPLTTSNMQTLFPQLVGQELQACNGT
FQFGISITGLEKLLNLNVQQLN*(1042)NSILSVTYHDYVTGETLTATTKLVALSTLSLVAKGAGV
VEFLLTAYPYTGN*(1093)ITFAPPWFIAENVVKQPFMTYSDLQFAKTNPSAILSLSTVN*(1134)ITVVGL
GGKASVYYNSTSGQTVITNIYGQTVATLSGNVLPTLTELAAGNGTFTGSLQFTIVPN*(1197)NTV
VQIPSSLTKTSFAVYTN*(1217)GSLAIVLNGKAYSLGPAGLFLLPFVTYTGSAIGAN*(1252)ATAIITVS
DGVGTSTTQVPITAEN*(1276)FTPIRLAPFQVPAQVPLPNAPKLKYEYN*(1304)GSIVITPQQQVLKIYV
TSILPYPQEFQIQAFVYEASQFNVHTGSPTAAPVYFSYSAVRAYPALGIGTSVPNLLVYV
QLQGISNLPAGKYVIVLSAVPFAGGPVLSEYPAQLIFTN*(1419)VTLTQ
|
| Sequence Around Glycosites (21 AA) | KLLNLNVQQLNNSILSVTYHD
FLLTAYPYTGNITFAPPWFIA
PSAILSLSTVNITVVGLGGKA
TGSLQFTIVPNNTVVQIPSSL
LTKTSFAVYTNGSLAIVLNGK
VTYTGSAIGANATAIITVSDG
STTQVPITAENFTPIRLAPFQ
PNAPKLKYEYNGSIVITPQQQ
SEYPAQLIFTNVTLTQ |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 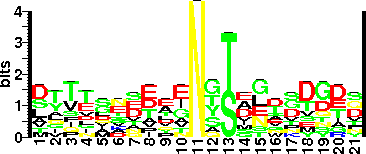 |
| Technique(s) used for Glycosylation Detection | Glycan identification by nano-LC-MS/MS |
| Technique(s) used for Glycosylated Residue(s) Detection | In-gel tryptic digestion and nano-LC-ES-MS/MS (nano-liquid chromatography-electrospray tandem mass spectrometry) |
| Protein Glycosylation- Implication | Glycosylation of S layer protein might contribute towards increased stability and half life of S layer structural proteins in extreme thermoacidic enviornments. |
| Glycan Information |
| Glycan Annotation | Linkage: β-GlcNAc-Asn.
N-linked hexasaccharide units - two residues each of Man and GlcNAc (one trisubstituted) and one residue each of Glc and 6-deoxy-6-sulfoglucose (6-sulfoquinovose, QuiS - a rare acidic sugar). β-D-Glcp-(1→4)-β-D-Quip(6--SO3-)-(1→3)-[α-D-Manp(1→4), α-D-Manp(1→6)] β-D-GlcpNAc-(1→4)-β-D-GlcpNAc-(1→Asn....
Microheterogeneity is observed as a family of glycans modify each of the glycosylation sites. The glycans are linked via a chitobiose core. |
| Technique(s) used for Glycan Identification | GC-MS of trimethylsilyl (TMS) methyl glycoside derivatives and nano-LC-ESMS analysis |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | Saci_1274 |
| OST NCBI Gene ID | 3473075 |
| OST GenBank Gene Sequence | 3473075 |
| OST Protein Name | Conserved membrane protein |
| OST UniProtKB/ SwissProt ID | Q4J9B4 |
| OST NCBI RefSeq | YP_255909.1 |
| OST EMBL-CDS | AAY80616.1 |
| OST UniProtKB Sequence | >tr|Q4J9B4|Q4J9B4_SULAC Conserved membrane protein OS=Sulfolobus acidocaldarius GN=Saci_1274 PE=4 SV=1
MQSTSILARIDKFKFLDAIIIGSLALFSILIRIISITAFPQTINGFDSWYLFYNALLIVK
AGGNWYAVPPDVHAWFPWGYFIELENTIGLPFLVALFSVPFYSIFGQNIVYTLTLVSPIV
LDGIGVVAAFLAVESITNSRVGGYIAAAITAFTPSLTYKNILGSLPKTSWGGVFVLFTIY
FLSLAIKKKKPLYGIPAGIMIFLANITWGGYTYIDISLAIAAFLIVLFNKNDEISAKTLT
ISGITAAFLTSLSPNTIGFMSEVAHGLALLIIPLFLYLDLYLRRVLPKDIVDSKNIVIGA
AIILLVSLVVLGSVAFKVQLIPSRYYAIINPFFQFTVPIDRTVAEYIPQSIAAMIQDFGI
GLFLSIIGIYFLLTRKQDMAGIWLVVLGAASIYGTSEQPYLFNYTIYIVAALAGVAVAEL
FSRFMERKIRIAPILMLTLIGVALLADAGIAVEASYAPQALINSSTSYLTTNYAWISALD
WINQNTPNNAFILSWWDYGYWIHAVGNRTVIDENNTLNGTQIKLMAEMFLNNESFAVNVL
ENDFHLYPYGNPNYTRPVYIVAYDAVTEYIVNNQYPVWFIGYPTNFPGTFIGYTTSLGDI
AKAIGAMTTIAGYNTNSYVNTTYINETASYAAQYNSQLASIIANSLPMAWTPKTYNSLIG
SMFIEAIQSLNQGPVQAPFSISLSQLLQSSSTSLYNPNALLPRVNLMYFKPVYIALFPLS
VTNALGGEAIVYIMVYIYQFVMPNVIIPPTISTA
|
| OST EC Number (BRENDA) | 2.4.1.119 |
| OST Genome Context | Q4J9B4 |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Peyfoon, E., Meyer, B., Hitchen, P.G., Panico, M., Morris, H.R., Haslam, S.M., Albers, S.V. and Dell, A. (2010) The S-layer glycoprotein of the crenarchaeote Sulfolobus acidocaldarius is glycosylated at multiple sites with chitobiose-linked N-glycans. Archaea, 2010. [PubMed: 20936123]
2) Veith, A., Klingl, A., Zolghadr, B., Lauber, K., Mentele, R., Lottspeich, F., Rachel, R., Albers, S.V. and Kletzin, A. (2009) Acidianus, Sulfolobus and Metallosphaera surface layers: structure, compositi |
| Additional Comments | Similar to eukaryotes and unlike other archaea, the tribranched glycan is linked via chitobiose core (GlcNAcβ1-4GlcNAc) to the Asn in S. acidocaldarius glycoproteins. Hence sulfolobus has been suggested as closest eukaryotic model for N glycosylation studies among archaeal speceis.
The glycosylation density at C terminus of the protein averages around 1 site per 30-40 residues.
Similarity found in glycan sequence in SlaA and cytochrome b558/566 subunit A strengthen the existing und |
| Year of Identification | 2010 |
| Year of Validation | 2010 |


