| ProGlyProt ID | AC120 |
| Organism Information |
| Organism Name | Sulfolobus acidocaldarius (DSM 639) |
| Domain | Archaea |
| Classification | Family: Sulfolobaceae
Order: Sulfolobales
Class: Thermoprotei or Crenarchaeota
Division or phylum: "Crenarchaeota" |
| Taxonomic ID (NCBI) | 2285 |
| Genome Sequence (s) |
| GeneBank | CP000077.1 |
| EMBL | CP000077 |
| Gene Information |
| Gene Name | cbsA(Saci_1858) |
| NCBI Gene ID | 3474781 |
| GenBank Gene Sequence | 3474781 |
| Protein Information |
| Protein Name | Cytochrome b558/566 subunit A (b type hemoprotein) |
| UniProtKB/SwissProt ID | O54088 |
| NCBI RefSeq | YP_256457.1 |
| EMBL-CDS | AAY81164.1 |
| UniProtKB Sequence | >sp|O54088|CBSA_SULAC Cytochrome b558/566 subunit A OS=Sulfolobus acidocaldarius GN=cbsA PE=1 SV=1
MSLKIKSKITIGVLLIIFLLSIIFTLENVSLAQTSPQISVYKVVGSADLSNPGSAGYWSQ
IPWTNISLTANIPMAPTSGLTHYLLVKAAWNGSWIFILEEWQAPEPAFNAWSTAVAGIYP
NASGPGLFRMIELTPGTTYSLERNYTNYVSIINGKEETGRIVFNYSGITLPAPNNTEITV
MSNGTILLWHSPRPVEDLLYNDGMFYGYYVNSTWYYPDRAAIMWYLGSGVPTKDDMNIGG
KYPGQQFDGITFKDAGGSLAQSGGSANIWMWVSGATWNNSTYDPAFKSNIWQNESLTGLS
YVDSGNHGFAVPLYTNNTNMYEVDTAGIWYTPVASEGLNGSLFFIWTGAKYENGSWVVEF
ARPLSVPLDYQPFMPNITVGKTYYVAFAVWQGRLGETLFDKSITSSFLSLELVTTPPTST
TTSTSPVTTISSAIPPVTLYVTIIGVVVALVALVILYVVFRR
|
| Sequence length | 462 AA |
| Subcellular Location | Integral membrane protein |
| Function | Membrane residing ectoenzyme. Due to the likely pseudoperiplasmic location, cytochrome c-like function is proposed analogous to the situation in Paracoccus denitrificans linking pseudoperiplasmic redox metabolism to membrane-residing electron transport systems. Cytochrome b558/566 is significantly up-regulated under certain organotrophic growth conditions in combination with low oxygen tension.
|
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked (O linked residues not yet identified) |
| Experimentally Validated Glycosite(s) in Full Length Protein | N144, N164 |
| Experimentally Validated Glycosite(s ) in Mature Protein | N144, N164 |
| Glycosite(s) Annotated Protein Sequence | >sp|O54088|CBSA_SULAC Cytochrome b558/566 subunit A OS=Sulfolobus acidocaldarius GN=cbsA PE=1 SV=1
MSLKIKSKITIGVLLIIFLLSIIFTLENVSLAQTSPQISVYKVVGSADLSNPGSAGYWSQ
IPWTNISLTANIPMAPTSGLTHYLLVKAAWNGSWIFILEEWQAPEPAFNAWSTAVAGIYP
NASGPGLFRMIELTPGTTYSLERN*(144)YTNYVSIINGKEETGRIVFN*(164)YSGITLPAPNNTEITV
MSNGTILLWHSPRPVEDLLYNDGMFYGYYVNSTWYYPDRAAIMWYLGSGVPTKDDMNIGG
KYPGQQFDGITFKDAGGSLAQSGGSANIWMWVSGATWNNSTYDPAFKSNIWQNESLTGLS
YVDSGNHGFAVPLYTNNTNMYEVDTAGIWYTPVASEGLNGSLFFIWTGAKYENGSWVVEF
ARPLSVPLDYQPFMPNITVGKTYYVAFAVWQGRLGETLFDKSITSSFLSLELVTTPPTST
TTSTSPVTTISSAIPPVTLYVTIIGVVVALVALVILYVVFRR
|
| Sequence Around Glycosites (21 AA) | TPGTTYSLERNYTNYVSIING
GKEETGRIVFNYSGITLPAPN
|
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 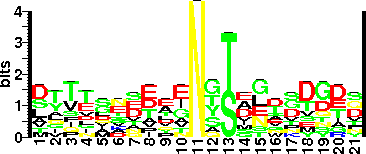 |
| Technique(s) used for Glycosylation Detection | Chemical deglycosylation (with TFMS) and mass shift on SDS-PAGE. |
| Technique(s) used for Glycosylated Residue(s) Detection | MALDI-MS, 1H-NMR spectroscopy of trypsin/pronase digested, anion-exchange HPLC purified glycan linked dipeptides. |
| Protein Glycosylation- Implication | The increased glycosylation of Cytochrome b558/566 subunit A has been suggested to have a role in the protection of enzyme against the harsh acidic conditions (pH 2 - 2.5) of S. acidocaldarius, in its thermoacidophilic habitat. Upon glycosylation with a low pKa sulfonate carrying glycans the protein achieves a negative surface potential at the locus of the cytochrome. The glycosylation of a major internal loop region also may improve resistance against hydrolytic cleavage which is even better th |
| Glycan Information |
| Glycan Annotation | Linkage: β-GlcNAc-Asn.
64-kDa glycoprotein expresses 17% glycosylation. Total sugar analysis revealed the relative proportions of approximately 7:2:2 for mannose, glucose and GlcNAc.
N-linked hexasaccharide units (at least seven)- two residues each of Man and GlcNAc (one trisubstituted) and one residue each of Glc and 6-deoxy-6-sulfoglucose (6-sulfoquinovose- a rare acidic sugar).
β-D-Glcp-(1→4)-β-D-Quip(6--SO3-)-(1→3)-[α-D-Manp(1→4), α-D-Manp(1→6)] β-D-GlcpNAc-(1→ |
| Technique(s) used for Glycan Identification | Identification of oligosaccharides using Gel Chromatography and GLC-MS (PNGase F treatment followed by HPAEC analysis). Glycan structure is elucidated with the help of 1H-NMR spectrum from two-dimensional 1H, 1H correlation NMR Spectroscopy . Linkage analysis was carried out using 13C NMR Chemical Shift analysis. |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | Saci_1274 |
| OST NCBI Gene ID | 3473075 |
| OST GenBank Gene Sequence | 3473075 |
| OST Protein Name | Conserved membrane protein |
| OST UniProtKB/ SwissProt ID | Q4J9B4 |
| OST NCBI RefSeq | YP_255909.1 |
| OST EMBL-CDS | AAY80616.1 |
| OST UniProtKB Sequence | >tr|Q4J9B4|Q4J9B4_SULAC Conserved membrane protein OS=Sulfolobus acidocaldarius GN=Saci_1274 PE=4 SV=1
MQSTSILARIDKFKFLDAIIIGSLALFSILIRIISITAFPQTINGFDSWYLFYNALLIVK
AGGNWYAVPPDVHAWFPWGYFIELENTIGLPFLVALFSVPFYSIFGQNIVYTLTLVSPIV
LDGIGVVAAFLAVESITNSRVGGYIAAAITAFTPSLTYKNILGSLPKTSWGGVFVLFTIY
FLSLAIKKKKPLYGIPAGIMIFLANITWGGYTYIDISLAIAAFLIVLFNKNDEISAKTLT
ISGITAAFLTSLSPNTIGFMSEVAHGLALLIIPLFLYLDLYLRRVLPKDIVDSKNIVIGA
AIILLVSLVVLGSVAFKVQLIPSRYYAIINPFFQFTVPIDRTVAEYIPQSIAAMIQDFGI
GLFLSIIGIYFLLTRKQDMAGIWLVVLGAASIYGTSEQPYLFNYTIYIVAALAGVAVAEL
FSRFMERKIRIAPILMLTLIGVALLADAGIAVEASYAPQALINSSTSYLTTNYAWISALD
WINQNTPNNAFILSWWDYGYWIHAVGNRTVIDENNTLNGTQIKLMAEMFLNNESFAVNVL
ENDFHLYPYGNPNYTRPVYIVAYDAVTEYIVNNQYPVWFIGYPTNFPGTFIGYTTSLGDI
AKAIGAMTTIAGYNTNSYVNTTYINETASYAAQYNSQLASIIANSLPMAWTPKTYNSLIG
SMFIEAIQSLNQGPVQAPFSISLSQLLQSSSTSLYNPNALLPRVNLMYFKPVYIALFPLS
VTNALGGEAIVYIMVYIYQFVMPNVIIPPTISTA
|
| OST EC Number (BRENDA) | 2.4.1.119 |
| OST Genome Context | Q4J9B4 |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Zahringer, U., Moll, H., Hettmann, T., Knirel, Y.A. and Schafer, G. (2000) Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius has a unique Asn-linked highly branched hexasaccharide chain containing 6-sulfoquinovose. Eur J Biochem, 267, 4144-4149. [PubMed: 10866817]
2) Hettmann, T., Schmidt, C.L., Anemuller, S., Zahringer, U., Moll, H., Petersen, A. and Schafer, G. (1998) Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius. A novel highly glycosylated, membrane-bo |
| Additional Comments | Provides first extensive study and a structure for oligosaccharide attached to a crenarchaeotal glycoprotein. The protein attached hexasaccharide is unique in terms of the presence of a trisubstituted N acetylglucosamine residue and a rare acidic residue Qui6S (sulfonated sugar).
Qui6S like sugar has previously been observed only in glycolipids of chloroplast and photosynthetic bacteria. Only other case where sufated sugar has been observed is H. salinaium.
Unusally, only one type of g |
| Year of Identification | 1998 |
| Year of Validation | 2000 |


