| ProGlyProt ID | AC117 |
| Organism Information |
| Organism Name | Methanothermus fervi |
| Domain | Archaea |
| Classification | Family: Methanothermaceae
Order: Methanobacteriales
Class: Methanobacteria or Archaeobacteria
Division or phylum: "Euryarchaeota" |
| Taxonomic ID (NCBI) | 2180 |
| Genome Sequence (s) |
| EMBL | X58297 |
| Gene Information |
| Gene Name | slgA |
| NCBI Gene ID | |
| Protein Information |
| Protein Name | SlgA (cell surface glycoprotein) |
| UniProtKB/SwissProt ID | P27373 |
| NCBI RefSeq | |
| EMBL-CDS | CAA41230.1 |
| UniProtKB Sequence | >sp|P27373|CSG_METFE Cell surface glycoprotein OS=Methanothermus fervidus GN=slgA PE=1 SV=1
MRKFTLLMLLLIVISMSGIAGAAEVKNLNTSKTFTKIQEAIDDPSTTDGNIIIVGPGNYT
ENILVNKSLTLKSNGSAIINAVSSEKSTITIKANNVWIEGFIIIGGKNGIYMENVTGCTI
TNNTIQNAFVSGWEYYGGNGICLVNSTNNTITNNIIRNNTWNGINVCESKGNIIKNNTIM
YSGGIGIYVWGFNKFEGNNIIENNRIINATYGGIYLFRPSNNKICRNYIANVSSGGGGMS
GAICIDVSDYNIVKDNIGINCDGGLFTDGMIGNEITNNIFKNCKVAVSESTYGPASRNNK
IYGNYFINYETAISDPKGELVDNIWNTTEGGNYWSNYTGNNTGDGTGNIPYYYDNKPLVV
DLAIEDIAAKPSGIEVRVKNLGKADIKKIDPLTKLKIKISCDNDVYETFIDPLSAGESQI
VRWDKIVPEGNHTIKAEIPYSAEGYLIGTNIRDADISNNVFSKIVQGFVQNKTFTITLTN
LGKSTITIKYYISIYTNPVNGTKVSYRELTITLKPNETKTIELGKYPFKYAVSGTMIVKN
PSRYRIPLNLRIKYEIEGLNPQMREISKYIAPRGEFRYIARYTGKEEGYADVW
|
| Sequence length | 593 AA |
| Subcellular Location | Surface |
| Function | In Archaea, which do not possess other cell wall components, the S-layer has to maintain the cell integrity and stabilize as well as to protect the cell against mechanical and osmotic stresses or extreme pH conditions. It is also predicted that the S-layer has to maintain or even determine the cell shape.
|
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | (Signal peptide: 1-22) N29 in case of precursor protein.
|
| Experimentally Validated Glycosite(s ) in Mature Protein | N7
|
| Glycosite(s) Annotated Protein Sequence | >sp|P27373|CSG_METFE Cell surface glycoprotein OS=Methanothermus fervidus GN=slgA PE=1 SV=1
MRKFTLLMLLLIVISMSGIAGAAEVKNLN*(29)TSKTFTKIQEAIDDPSTTDGNIIIVGPGNYT
ENILVNKSLTLKSNGSAIINAVSSEKSTITIKANNVWIEGFIIIGGKNGIYMENVTGCTI
TNNTIQNAFVSGWEYYGGNGICLVNSTNNTITNNIIRNNTWNGINVCESKGNIIKNNTIM
YSGGIGIYVWGFNKFEGNNIIENNRIINATYGGIYLFRPSNNKICRNYIANVSSGGGGMS
GAICIDVSDYNIVKDNIGINCDGGLFTDGMIGNEITNNIFKNCKVAVSESTYGPASRNNK
IYGNYFINYETAISDPKGELVDNIWNTTEGGNYWSNYTGNNTGDGTGNIPYYYDNKPLVV
DLAIEDIAAKPSGIEVRVKNLGKADIKKIDPLTKLKIKISCDNDVYETFIDPLSAGESQI
VRWDKIVPEGNHTIKAEIPYSAEGYLIGTNIRDADISNNVFSKIVQGFVQNKTFTITLTN
LGKSTITIKYYISIYTNPVNGTKVSYRELTITLKPNETKTIELGKYPFKYAVSGTMIVKN
PSRYRIPLNLRIKYEIEGLNPQMREISKYIAPRGEFRYIARYTGKEEGYADVW
|
| Sequence Around Glycosites (21 AA) | IAGAAEVKNLNTSKTFTKIQE |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 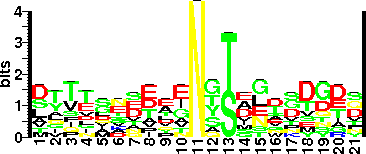 |
| Technique(s) used for Glycosylation Detection | Carbohydrate detection (TLC, thin layer chromatography) |
| Technique(s) used for Glycosylated Residue(s) Detection | Edman degradation |
| Protein Glycosylation- Implication | |
| Glycan Information |
| Glycan Annotation | Linkage: β-GlcNAc-Asn.
N-linked heterosaccharide is 17 % of molecular weight of the protein. Composition of the hexasaccharide (1061.83 Da) is α-D-3-O-MetManp-(1→6)-α-D-3-O-MetManp-[(1→2)-α-D-Manp]3-(1→4)-D-GalNAc.
3-O-methylglucose, N-acetylglucosamine, and galactose are also found in lesser amounts. 3-O-methylmannose could be partly replaced by 3-O-methylglucose.
|
| Technique(s) used for Glycan Identification | Methylation analysis, plasma desorption mass spectrometry, and high field 1H, 13C NMR spectroscopy- COSY (correlated spectroscopy), 1D-, 2D-TOCSY (total correlation spectroscopy), and HMBC (heteronuclear multiple bond coherence), HMQC (heteronuclear multiple quantum correlation). |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | Mfer_0177 |
| OST NCBI Gene ID | 9961890 |
| OST GenBank Gene Sequence | 9961890 |
| OST Protein Name | Oligosaccharyl transferase stt3 subunit |
| OST UniProtKB/ SwissProt ID | E3GXE8 |
| OST NCBI RefSeq | YP_004003742.1 |
| OST EMBL-CDS | ADP76980.1 |
| OST UniProtKB Sequence | >tr|E3GXE8|E3GXE8_METFE Oligosaccharyl transferase STT3 subunit OS=Methanothermus fervidus DSM 2088 GN=Mfer_0177 PE=4 SV=1
MKKKMLNTIKQIAIIAILFSLAFYIRSEAVNIGGVPPDQKNFYKDPSTGLPYFSEIDSYY
NLRLTTNFLTKGMLGDTKVNGKDWDLHSYYPPGRPVDYPPMIVYVTSFLYKFVNIFKKVS
LPVVAFWTGAFIGSLVVIPAYLFVRKITNDYGGITAALLVAFAPVYFAHTFAGLFRTAMF
NVLFPILIMWFFAESIVAKTFKRRVIFAVATAISIILFSLSWVGYVFYLAVIFVVALVYF
GLRYFIERGKVEYVSKLDWIKGQRELLALLIVAIICGGILTIGGGPSAIPNALSQLVGAV
QMQALTQTSSYPNVYISVAELQVPPFVMPIPGKPGSVFLPNQMSAVGQIGGLGVFLLGVA
GIIALLWKAGSIYRSEFSLTKDFLKTTPKIRAGKTSKFKKRKRRISEEVSRDIQIEHLFY
AVLFTIWSIMAGYAVTKGIRFAEPFSIPMSLSAGIFVGFAADFVKKHVRAELTALTLAFI
CGILAASPFAPTVQMKFLAGIISAILVYIVRKHSLRSVIIALLIVGVAATPSIYGAQQFA
SQIVPSSNTGMYNATKWIKDHTTPGTVIMSWWDFGHFFTYEADRPVTFDGGSQNTPRAYW
IGKALLTNNETLSAGILRMLASSGDKAYLTLDNYTHDTAKTVEILDKTLGVPKSEARNIM
INEYKLTPQQADTILQYTHPDKKRPFVLVCSSDMIPKAAWWSYFGSWNFETKNGTHYFYH
ISQQVSPPQKEKDKTVIYTINEVMQNQMICTKIEKKANEINATILMGTKGNLNEVKPHKL
VIIEGNRVVKNEIVDKDSNYGLVVIGQGGSYVTIVMDKQLEDSVFTKLFILGGFNQTTFK
YLHSEPGVSIWTAQ |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Karcher, U., Schroder, H., Haslinger, E., Allmaier, G., Schreiner, R., Wieland, F., Haselbeck, A. and Konig, H. (1993) Primary structure of the heterosaccharide of the surface glycoprotein of Methanothermus fervidus. J Biol Chem, 268, 26821-26826. [PubMed: 8262914]
2) Kalmokoff, M.L., Koval, S.F. and Jarrell, K.F. (1992) Relatedness of the flagellins from methanogens. Arch Microbiol, 157, 481-487. [PubMed: 1503530]
3) Brockl, G., Behr, M., Fabry, S., Hensel, R., Kaudewitz, H., Biend |
| Additional Comments | |
| Year of Identification | 1989 |
| Year of Validation | 1991 |


