| ProGlyProt ID | AC116 |
| Organism Information |
| Organism Name | Methanococcus voltae |
| Domain | Archaea |
| Classification | Family: Methanococcaceae
Order: Methanococcales
Class: Methanococci or Methanothermea
Division or phylum: "Euryarchaeota" |
| Taxonomic ID (NCBI) | 2188 |
| Genome Sequence (s) |
| EMBL | M59200 |
| Gene Information |
| Gene Name | sla |
| NCBI Gene ID | |
| Protein Information |
| Protein Name | S-layer protein |
| UniProtKB/SwissProt ID | Q50833 |
| NCBI RefSeq | |
| EMBL-CDS | AAA93515.1 |
| UniProtKB Sequence | >sp|Q50833|CSG_METVO S-layer protein (Fragment) OS=Methanococcus voltae GN=sla PE=1 SV=2
KKIGAIAAGSAMVASALATGVFAVEKIGDVEGFKVIDNGEPTADIVVGSTAAAADVVSAA
NVAAKVGSMMFKEGEAASGSAKLTVKASAESDDANLKSLLTNGTNDFTELDAGKEAFVVA
AADSDYSDALINATTGFANIADNVLYDQAKLAAAVSLGDLSTLSVVKDIDPSDWYADKNK
AADVATKDYYDQDGDAVEMLMATVASNDDGKSLTVDEDGVLYASIAYDDDNEDFQRATQV
LKEGNRLPFLGEEYALVKLDTDDDIVYLGKEVFDGVLKEGDTYNIGDGYELKVVAILKSG
DEYKISLQLMKDGKVVAEKFDKVSATSALKMIYTPGNIGIVVNEAWENVGQDYGYGSTLI
TKDVIALELGEEYIPDWEVVTIEKDTTTDNTKDSKMTLSDDKITKDNTYGIGLQYVGDEE
DNFKSGKAIKIAKYAELELDDEDKEDTKLNLFFSMDETKEATLAAGQKVTVLNSDITLSE
VMADAKAPVAFKAPLAVLDTEVSLDAANKKLILVGGPVANALTKELADAGKIEMTVESPA
TLAVVAGAANGNDVLVVAGGDRAATAEAANALIEML
|
| Sequence length | 576 AA |
| Subcellular Location | Surface |
| Function | In Archaea, which do not possess other cell wall components, the S-layer has to maintain the cell integrity and stabilize as well as to protect the cell against mechanical and osmotic stresses or extreme pH conditions. It is also predicted that the S-layer has to maintain or even determine the cell shape.
|
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | (Signal peptide: 1-23) N102, also N132 in a second version of this strain (M. voltae PS*). |
| Experimentally Validated Glycosite(s ) in Mature Protein | N79, also N109 in a second version of this strain (M. voltae PS*). |
| Glycosite(s) Annotated Protein Sequence | >sp|Q50833|CSG_METVO S-layer protein (Fragment) OS=Methanococcus voltae GN=sla PE=1 SV=2
KKIGAIAAGSAMVASALATGVFAVEKIGDVEGFKVIDNGEPTADIVVGSTAAAADVVSAA
NVAAKVGSMMFKEGEAASGSAKLTVKASAESDDANLKSLLTN*(102)GTNDFTELDAGKEAFVVA
AADSDYSDALINATTGFANIADNVLYDQAKLAAAVSLGDLSTLSVVKDIDPSDWYADKNK
AADVATKDYYDQDGDAVEMLMATVASNDDGKSLTVDEDGVLYASIAYDDDNEDFQRATQV
LKEGNRLPFLGEEYALVKLDTDDDIVYLGKEVFDGVLKEGDTYNIGDGYELKVVAILKSG
DEYKISLQLMKDGKVVAEKFDKVSATSALKMIYTPGNIGIVVNEAWENVGQDYGYGSTLI
TKDVIALELGEEYIPDWEVVTIEKDTTTDNTKDSKMTLSDDKITKDNTYGIGLQYVGDEE
DNFKSGKAIKIAKYAELELDDEDKEDTKLNLFFSMDETKEATLAAGQKVTVLNSDITLSE
VMADAKAPVAFKAPLAVLDTEVSLDAANKKLILVGGPVANALTKELADAGKIEMTVESPA
TLAVVAGAANGNDVLVVAGGDRAATAEAANALIEML
|
| Sequence Around Glycosites (21 AA) | DDANLKSLLTNGTNDFTELDA
ADSDYSDALINATTGFANIAD
|
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 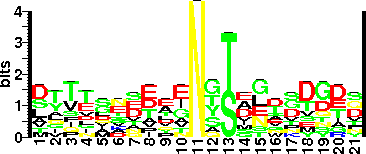 |
| Technique(s) used for Glycosylation Detection | Glycan (779 Da) identification by nano-LC-MS/MS |
| Technique(s) used for Glycosylated Residue(s) Detection | Nano-LC-MS/MS (nano-liquid chromatography-electrospray tandem mass spectrometry) |
| Protein Glycosylation- Implication | Mutations that ultimately alter the structure of the S-layer protein could affect the protein interaction and disrupt the structural integrity of the cell. As a result, such mutants could suffer serious cell stability problems. It is this effect on the S-layer that likely explains why the agl mutants are so unstable. |
| Glycan Information |
| Glycan Annotation | Linkage: β-GlcNAc-Asn.
Trisaccharide (779 Da) composed of β-Manp NAcA6Thr-(1– 4)- β-Glcp NAc3NAcA-(1–3)-β-Glcp NAc. The sugars are mannuronic acid with the attached threonine, diacetylated glucuronic acid, N-acetylglucosamine.
In a second version of this strain (M. voltae PS*), the glycan is modified with one additional residue (either 220 or 262 Da) linked to the terminal modified mannuronic acid. |
| Technique(s) used for Glycan Identification | NanoLC–MS/MS analysis and NMR spectroscopy- COSY (correlated spectroscopy), TOCSY (total correlation spectroscopy), NOESY (nuclear Overhauser effect spectroscopy) spectra, and 1H-13C HMBC (heteronuclear multiple bond coherence) spectra. |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | aglB |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | AglB |
| OST UniProtKB/ SwissProt ID | Q2EMT4 |
| OST NCBI RefSeq | |
| OST EMBL-CDS | Â ABD17750.1 |
| OST UniProtKB Sequence | >tr|Q2EMT4|Q2EMT4_METVO STT3 OS=Methanococcus voltae GN=MVO1749 PE=4 SV=1
MTENNEKVKNSDSANNQSSKNSKFNFNFEDKKVKCAKTILIIIFLAFLSFQMRAQTADMG
FTTNEQYLDVFSDDNGRMYLTALDPYYYLRMSENYLENGHTGDTLKNIDGQQVPWDSYKY
GPTGARATFNLLSVVTVWVYQVWHAMDSTVTLMNAAFWVPAILSMFLITPIFFTVRRITS
SDIGGAVAAILASLSPSIFVKTVAGFSDTPILEILPLLFIVWFIIEAIHYSKEKNYKSLI
YGLLATLMLALYPFMWSAWWYGYYIVIAFLVIYAIYKGISYNSIAKYTKSKNNNHKDKIE
SEKLEMLNILKISGLFIIGGAVLITALYGVSTTMNALQAPLNYLGLDEVSSQTGWPNVLT
TVSELDTASLDEIISSSLGSIHLFAIGLIGIFLSLFRKVLTPVKQISNGLAEKLDIKYAL
LLIIWFAVTFLAASKGVRFVALMVPPLSIGVGIFVGFIEQFIKNNLDKKYEYVAYPTIAI
IVLYALFTIYRADSADLVRMLLPSNYVPIAEGIMLASLAVLIIYKVAELIAESNKKLVMN
KIFMILLAIGLITPTIATIVPFYSVPTYNDGWGESLEWINTQTPNNSVVTCWWDNGHIYT
WKTDRMVTFDGSSQNTPRAYWVGRAFSTSNESLANGIFRMLASSGDKAYTTDSVLIKKTG
SIKNTVDVLNEILPLTKSDAQKALKNSSYKFTDTEVSEILDATHPKVTNPDYLITYNRMT
SIASVWSYFGNWDFNLPAGTSRSEREAGSFQGLQTYATNINDTLIVRSLIQQTAEYNIYT
LIEVRNETLTGAMMAVTNDGQMQTQQLNMHKVKLMVNENGKSKMYNSLADPDGQLSLLIK
VDKNSIIGTDGSNNPVYSSSSWMATANLEDSVYSKLHFFDGEGLDTIKLEKESLDPTANG
VQPGFKVFSVD |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | AglH, AglC, AglK, AglA glycosyltransferases are involved in the biosynthesis of the glycan. AglH is a GlcNAc-1-phosphate transferase transfering first sugar to dolichol pyrophosphate. AglC and AglK add the second sugar residue and AglA adds the third sugar to the glycan.
Characterized by gene deletion studies |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Chaban, B., Logan, S.M., Kelly, J.F. and Jarrell, K.F. (2009) AglC and AglK are involved in biosynthesis and attachment of diacetylated glucuronic acid to the N-glycan in Methanococcus voltae. J Bacteriol, 191, 187-195. [PubMed: 18978056]
2) Shams-Eldin, H., Chaban, B., Niehus, S., Schwarz, R.T. and Jarrell, K.F. (2008) Identification of the archaeal alg7 gene homolog (encoding N-acetylglucosamine-1-phosphate transferase) of the N-linked glycosylation system by cross-domain complementati |
| Additional Comments | |
| Year of Identification | 2005 |
| Year of Validation | 2005 |


