| ProGlyProt ID | AC112 |
| Organism Information |
| Organism Name | Methanococcus voltae PS |
| Domain | Archaea |
| Classification | Family: Methanococcaceae
Order: Methanococcales
Class: Methanococci or Methanothermea
Division or phylum: "Euryarchaeota" |
| Taxonomic ID (NCBI) | 2188 |
| Genome Sequence (s) |
| EMBL | M72148 |
| Gene Information |
| Gene Name | flaB1 |
| NCBI Gene ID | |
| Protein Information |
| Protein Name | Fla B1 |
| UniProtKB/SwissProt ID | P27803 |
| NCBI RefSeq | |
| EMBL-CDS | AAA73074.1 |
| UniProtKB Sequence | >sp|P27803|FLAB1_METVO Flagellin B1 OS=Methanococcus voltae GN=flaB1 PE=1 SV=1
MNIKEFLSNKKGASGIGTLIVFIAMVLVAAVAASVLINTSGFLQQKASTTGKESTEQVAS
GLQVTGVTGVNSTDSENITHIVAYITPKAGSSAIDLSQAKLFVTYNGVSAVLKANESAID
GTSGTPDVLSATLLSNTSATEYQVVSLQDFDGSVAKNQVINKGDLVAIRISTGQVFSSTK
GIPTRAHVSGKLQPEFGAPGIIEFTTPATFTTKVVELQ
|
| Sequence length | 218 AA |
| Subcellular Location | Surface associated |
| Function | Flagellin structural protein |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | (Propeptide: 1-12) N38, N71, N77, N115, N136
|
| Experimentally Validated Glycosite(s ) in Mature Protein | N26, N59, N65, N103, N124
|
| Glycosite(s) Annotated Protein Sequence | >sp|P27803|FLAB1_METVO Flagellin B1 OS=Methanococcus voltae GN=flaB1 PE=1 SV=1
MNIKEFLSNKKGASGIGTLIVFIAMVLVAAVAASVLIN*(38)TSGFLQQKASTTGKESTEQVAS
GLQVTGVTGVN*(71)STDSEN*(77)ITHIVAYITPKAGSSAIDLSQAKLFVTYNGVSAVLKAN*(115)ESAID
GTSGTPDVLSATLLSN*(136)TSATEYQVVSLQDFDGSVAKNQVINKGDLVAIRISTGQVFSSTK
GIPTRAHVSGKLQPEFGAPGIIEFTTPATFTTKVVELQ
|
| Sequence Around Glycosites (21 AA) | VAAVAASVLINTSGFLQQKAS
GLQVTGVTGVNSTDSENITHI
VTGVNSTDSENITHIVAYITP
YNGVSAVLKANESAIDGTSGT
PDVLSATLLSNTSATEYQVVS
|
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 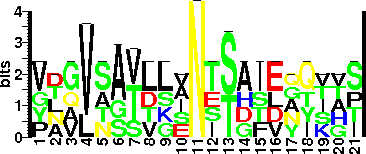 |
| Technique(s) used for Glycosylation Detection | Mass shift detected on SDS-polyacrylamide gel and glycan identification by nano-LC-MS/MS |
| Technique(s) used for Glycosylated Residue(s) Detection | Nano-LC-MS/MS (nano-liquid chromatography-electrospray tandem mass spectrometry) |
| Protein Glycosylation- Implication | The glycans on flagellin proteins are involved in the flagella assembly process. |
| Glycan Information |
| Glycan Annotation | Linkage: β-GlcNAc-Asn.
Trisaccharide (779 Da) composed of β-Manp NAcA6Thr-(1– 4)- β-Glcp NAc3NAcA-(1–3)-β-Glcp NAc. The sugars are mannuronic acid with the attached threonine, diacetylated glucuronic acid, N-acetylglucosamine.
In a second version of this strain (M. voltae PS*), the glycan is modified with one additional residue (either 220 or 262 Da) linked to the terminal modified mannuronic acid. |
| Technique(s) used for Glycan Identification | NanoLC–MS/MS analysis and NMR spectroscopy- COSY (correlated spectroscopy), TOCSY (total correlation spectroscopy), NOESY (nuclear Overhauser effect spectroscopy) spectra, and 1H-13C HMBC (heteronuclear multiple bond coherence) spectra. |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | aglB |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | AglB |
| OST UniProtKB/ SwissProt ID | Q2EMT4 |
| OST NCBI RefSeq | |
| OST EMBL-CDS | Â ABD17750.1 |
| OST UniProtKB Sequence | >tr|Q2EMT4|Q2EMT4_METVO STT3 OS=Methanococcus voltae GN=MVO1749 PE=4 SV=1
MTENNEKVKNSDSANNQSSKNSKFNFNFEDKKVKCAKTILIIIFLAFLSFQMRAQTADMG
FTTNEQYLDVFSDDNGRMYLTALDPYYYLRMSENYLENGHTGDTLKNIDGQQVPWDSYKY
GPTGARATFNLLSVVTVWVYQVWHAMDSTVTLMNAAFWVPAILSMFLITPIFFTVRRITS
SDIGGAVAAILASLSPSIFVKTVAGFSDTPILEILPLLFIVWFIIEAIHYSKEKNYKSLI
YGLLATLMLALYPFMWSAWWYGYYIVIAFLVIYAIYKGISYNSIAKYTKSKNNNHKDKIE
SEKLEMLNILKISGLFIIGGAVLITALYGVSTTMNALQAPLNYLGLDEVSSQTGWPNVLT
TVSELDTASLDEIISSSLGSIHLFAIGLIGIFLSLFRKVLTPVKQISNGLAEKLDIKYAL
LLIIWFAVTFLAASKGVRFVALMVPPLSIGVGIFVGFIEQFIKNNLDKKYEYVAYPTIAI
IVLYALFTIYRADSADLVRMLLPSNYVPIAEGIMLASLAVLIIYKVAELIAESNKKLVMN
KIFMILLAIGLITPTIATIVPFYSVPTYNDGWGESLEWINTQTPNNSVVTCWWDNGHIYT
WKTDRMVTFDGSSQNTPRAYWVGRAFSTSNESLANGIFRMLASSGDKAYTTDSVLIKKTG
SIKNTVDVLNEILPLTKSDAQKALKNSSYKFTDTEVSEILDATHPKVTNPDYLITYNRMT
SIASVWSYFGNWDFNLPAGTSRSEREAGSFQGLQTYATNINDTLIVRSLIQQTAEYNIYT
LIEVRNETLTGAMMAVTNDGQMQTQQLNMHKVKLMVNENGKSKMYNSLADPDGQLSLLIK
VDKNSIIGTDGSNNPVYSSSSWMATANLEDSVYSKLHFFDGEGLDTIKLEKESLDPTANG
VQPGFKVFSVD |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | AglH, AglC, AglK, AglA glycosyltransferases are involved in the biosynthesis of the glycan. AglH is a GlcNAc-1-phosphate transferase transfering first sugar to dolichol pyrophosphate. AglC and AglK add the second sugar residue and AglA adds the third sugar to the glycan.
Characterized by gene deletion studies |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Chaban, B., Logan, S.M., Kelly, J.F. and Jarrell, K.F. (2009) AglC and AglK are involved in biosynthesis and attachment of diacetylated glucuronic acid to the N-glycan in Methanococcus voltae. J Bacteriol, 191, 187-195. [PubMed: 18978056]
2) Shams-Eldin, H., Chaban, B., Niehus, S., Schwarz, R.T. and Jarrell, K.F. (2008) Identification of the archaeal alg7 gene homolog (encoding N-acetylglucosamine-1-phosphate transferase) of the N-linked glycosylation system by cross-domain complementati |
| Additional Comments | Post translational modification was detected in the year 1999. It was confirmed as glycosylation (by identifying the glycans) in 2005. |
| Year of Identification | 2005 |
| Year of Validation | 2005 |


