| ProGlyProt ID | AC105 |
| Organism Information |
| Organism Name | Halobacterium salinarum (halobium) R1M1/NRC-1 |
| Domain | Archaea |
| Classification | Family: Halobacteriaceae
Order: Halobacteriales
Class: Halobacteria or Halomebacteria
Division or phylum: "Euryarchaeota" |
| Taxonomic ID (NCBI) | 64091 |
| Genome Sequence (s) |
| GeneBank | AE004437.1 |
| EMBL | AE004437 |
| Gene Information |
| Gene Name | flaB1 (VNG_0960G) |
| NCBI Gene ID | 1447685 |
| GenBank Gene Sequence | 1447685 |
| Protein Information |
| Protein Name | Flagellin B1 |
| UniProtKB/SwissProt ID | P61116 |
| NCBI RefSeq | NP_279903.1 |
| EMBL-CDS | AAG19383.1 |
| UniProtKB Sequence | >sp|P61116|FLAB1_HALSA Flagellin B1 OS=Halobacterium salinarium (strain ATCC 700922 / JCM 11081 / NRC-1) GN=flaB1 PE=3 SV=1
MFEFITDEDERGQVGIGTLIVFIAMVLVAAIAAGVLINTAGYLQSKGSATGEEASAQVSN
RINIVSAYGNVNNEKVDYVNLTVRQAAGADNINLTKSTIQWIGPDRATTLTYSSNSPSSL
GENFTTESIKGSSADVLVDQSDRIKVIMYASGVSSNLGAGDEVQLTVTTQYGSKTTYWAQ
VPESLKDKNAVTL |
| Sequence length | 193 AA |
| Subcellular Location | Flagellum |
| Function | Flagellin is the structural subunit of the flagellar filaments. |
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked |
| Experimentally Validated Glycosite(s) in Full Length Protein | N93 |
| Experimentally Validated Glycosite(s ) in Mature Protein | N93 |
| Glycosite(s) Annotated Protein Sequence | >sp|P61116|FLAB1_HALSA Flagellin B1 OS=Halobacterium salinarium (strain ATCC 700922 / JCM 11081 / NRC-1) GN=flaB1 PE=3 SV=1
MFEFITDEDERGQVGIGTLIVFIAMVLVAAIAAGVLINTAGYLQSKGSATGEEASAQVSN
RINIVSAYGNVNNEKVDYVNLTVRQAAGADNIN*(93)LTKSTIQWIGPDRATTLTYSSNSPSSL
GENFTTESIKGSSADVLVDQSDRIKVIMYASGVSSNLGAGDEVQLTVTTQYGSKTTYWAQ
VPESLKDKNAVTL |
| Sequence Around Glycosites (21 AA) | VRQAAGADNINLTKSTIQWIG |
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 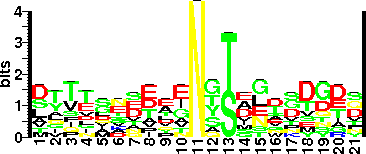 |
| Technique(s) used for Glycosylation Detection | Mass shift on SDS-PAGE after deglycosylation with anhydrous hydrogen fluoride. |
| Technique(s) used for Glycosylated Residue(s) Detection | Glycopeptide sequencing |
| Protein Glycosylation- Implication | |
| Glycan Information |
| Glycan Annotation | Linkage: Glc-Asn.
Sulfated oligosaccharides that resemble those of the cell-suface glycoprotein in the bacterium are present. Glucose and glucuronic acid are the constituents of the glycans which are of the type GlcA-(1→4)-GlcA-(1→4)-GlcA-(1→4)-Glc. |
| Technique(s) used for Glycan Identification | GLC-MS (gas liquid chromatography-mass spectrometry) after perfluoropropionylation. |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | |
| OST NCBI Gene ID | |
| OST GenBank Gene Sequence | |
| OST Protein Name | |
| OST UniProtKB/ SwissProt ID | |
| OST NCBI RefSeq | |
| OST EMBL-CDS | |
| OST UniProtKB Sequence | |
| OST EC Number (BRENDA) | |
| OST Genome Context | |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Cohen-Krausz, S. and Trachtenberg, S. (2002) The structure of the archeabacterial flagellar filament of the extreme halophile Halobacterium salinarum R1M1 and its relation to eubacterial flagellar filaments and type IV pili. J Mol Biol, 321, 383-395. [PubMed: 12162953]
2) Gerl, L., Deutzmann, R. and Sumper, M. (1989) Halobacterial flagellins are encoded by a multigene family. Identification of all five gene products. FEBS Lett, 244, 137-140. [PubMed: 2924901]
3) Gerl, L. and Sumper, M |
| Additional Comments | Removal of Mg++ ions in the growth medium inhibits glycosylation of proteins in vivo. This leads to the reduction in the molecular masses of newly synthesized flagellins. There might be another modification of the flagellins that is resistant to HF treatment. Halobacterial glycoproteins including flagellins are glycosylated at the extracellular surface of the cell membrane. |
| Year of Identification | 1985 |
| Year of Validation | 1988 |


