| ProGlyProt ID | AC102 |
| Organism Information |
| Organism Name | Halobacterium salinarum (halobium) R1M1/NRC-1 |
| Domain | Archaea |
| Classification | Family: Halobacteriaceae
Order: Halobacteriales
Class: Halobacteria or Halomebacteria
Division or phylum: "Euryarchaeota" |
| Taxonomic ID (NCBI) | 64091 |
| Genome Sequence (s) |
| GeneBank | AE004437.1 |
| EMBL | AE004437 |
| Gene Information |
| Gene Name | csg (VNG_2679G) |
| NCBI Gene ID | 1449008 |
| GenBank Gene Sequence | 1449008 |
| Protein Information |
| Protein Name | S layer glycoprotein |
| UniProtKB/SwissProt ID | Q9HM69 |
| NCBI RefSeq | NP_281222.1 |
| EMBL-CDS | AAG20702.1 |
| UniProtKB Sequence | >sp|Q9HM69|CSG_HALSA Cell surface glycoprotein OS=Halobacterium salinarium GN=csg PE=1 SV=2
MTDTTGKLRAVLLTALMVGSVIGAGVAFTGGAAAANASDLNDYQRFNENTNYTYSTASED
GKTEGSVASGATIFQGEEDVTFRKLDNEKEVSPATLSRTGGSDEGVPLQMPIPEDQSTGS
YDSNGPDNDEADFGVTVQSPSVTMLEVRNNADNDVTGGVLNTQQDESSIAVDYNYYAAED
LELTVEDEDGLDVTDEILAADQSGGAYEDGTGNNGPNTLRFDIDPNNVDAGDYTVSVEGV
EDLDFGDATESASVTISSSNKASLNLAEDEVVQGANLKYTIENSPEGNYHAVTIDSSDFR
DSSSGADAAKVMRSVGDTVDTGLVVDNDSTTEIVDDYENTSISDVDYAYAIVEIDDGNGV
GSIETQYLDDSSADIDLYPASDTEDAPDYVNSNEELTNGSALDGVSTDDDTDFDVTQGDI
TLDNPTGAYVVGSEVDINGTANEGTDDVVLYARDNNDFELVTVDGEKSIEVDSDDTFEEE
DITLSDGDKGGDDILGLPGTYRLGIIAKSDAVNSSGGVKDNIDTSDFNQGVSSTSSIRVT
DTELTASFETYNGQVADDDNQIDVEGTAPGKDNVAAIIIGSRGKVKFQSISVDSDDTFDE
EDIDISELRQGSASAHILSSGRDGKFGEDTANSISDLEDEVGNYTSGSPTGDQIRDRILS
NTVDDTASDDLIVTQQFRLVDGLTTIEATEGGEAGGSLTVMGTTNRKADDNTITVELLQG
DASIEINSTDEWNSDGQWSVDVPLSNVEPGNYTVEADDGDNTDRQNVEIVEELEEPDQTT
VDQPENNQTMTTTMTETTTETTTEMTTTQENTTENGSEGTSDGESGGSIPGFGVGVALVA
VLGAALLALRQN
|
| Sequence length | 852 AA |
| Subcellular Location | Surface |
| Function | In Archaea, which do not possess other cell wall components, the S-layer has been implicated in the maintaineance of the cell integrity and stabilize as well as to protect the cell against mechanical and osmotic stresses or extreme pH conditions. It is also predicted that the S-layer has to maintain or even determine the cell shape.
|
| Protein Structure |
| PDB ID | |
| Glycosylation Status |
| Glycosylation Type | N (Asn) linked (O, Thr-linked residues not known) |
| Experimentally Validated Glycosite(s) in Full Length Protein | (Signal peptide: 1-34) N36, N339, N398, N438, N513, N643, N727, N751, N787, N811, N815 (N36, N513 and N643 were confirmed glycosylated directly by glycopeptide sequence analysis , the reference no. 2 mentions that ten sulfated saccharides are N-linked to the protein imlplying that most or all sites are glycosylated.) |
| Experimentally Validated Glycosite(s ) in Mature Protein | N2, N305, N364, N404, N479, N609, N693, N717, N753, N777, N781 |
| Glycosite(s) Annotated Protein Sequence | >sp|Q9HM69|CSG_HALSA Cell surface glycoprotein OS=Halobacterium salinarium GN=csg PE=1 SV=2
MTDTTGKLRAVLLTALMVGSVIGAGVAFTGGAAAAN*(36)ASDLNDYQRFNENTNYTYSTASED
GKTEGSVASGATIFQGEEDVTFRKLDNEKEVSPATLSRTGGSDEGVPLQMPIPEDQSTGS
YDSNGPDNDEADFGVTVQSPSVTMLEVRNNADNDVTGGVLNTQQDESSIAVDYNYYAAED
LELTVEDEDGLDVTDEILAADQSGGAYEDGTGNNGPNTLRFDIDPNNVDAGDYTVSVEGV
EDLDFGDATESASVTISSSNKASLNLAEDEVVQGANLKYTIENSPEGNYHAVTIDSSDFR
DSSSGADAAKVMRSVGDTVDTGLVVDNDSTTEIVDDYEN*(339)TSISDVDYAYAIVEIDDGNGV
GSIETQYLDDSSADIDLYPASDTEDAPDYVNSNEELTN*(398)GSALDGVSTDDDTDFDVTQGDI
TLDNPTGAYVVGSEVDIN*(438)GTANEGTDDVVLYARDNNDFELVTVDGEKSIEVDSDDTFEEE
DITLSDGDKGGDDILGLPGTYRLGIIAKSDAVN*(513)SSGGVKDNIDTSDFNQGVSSTSSIRVT
DTELTASFETYNGQVADDDNQIDVEGTAPGKDNVAAIIIGSRGKVKFQSISVDSDDTFDE
EDIDISELRQGSASAHILSSGRDGKFGEDTANSISDLEDEVGN*(643) YTSGSPTGDQIRDRILS
NTVDDTASDDLIVTQQFRLVDGLTTIEATEGGEAGGSLTVMGTTNRKADDNTITVELLQG
DASIEIN*(727)STDEWNSDGQWSVDVPLSNVEPGN*(751)YTVEADDGDNTDRQNVEIVEELEEPDQTT
VDQPENN*(787)QTMTTTMTETTTETTTEMTTTQEN*(811)TTEN*(815)GSEGTSDGESGGSIPGFGVGVALVA
VLGAALLALRQN
|
| Sequence Around Glycosites (21 AA) | VAFTGGAAAANASDLNDYQRF
STTEIVDDYENTSISDVDYAY
DYVNSNEELTNGSALDGVSTD
AYVVGSEVDINGTANEGTDDV
LGIIAKSDAVNSSGGVKDNID
SISDLEDEVGNYTSGSPTGDQ
LLQGDASIEINSTDEWNSDGQ
DVPLSNVEPGNYTVEADDGDN
DQTTVDQPENNQTMTTTMTET
TTTEMTTTQENTTENGSEGTS
MTTTQENTTENGSEGTSDGES
|
| Glycosite Sequence Logo | seqlogo |
| Glycosite Sequence Logo | 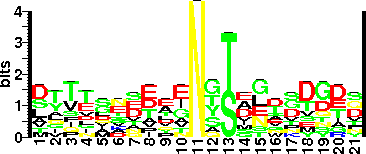 |
| Technique(s) used for Glycosylation Detection | Periodate-arsenite-Schiff reagent staining and carbohydrate analysis using GC. |
| Technique(s) used for Glycosylated Residue(s) Detection | Glycopeptide sequencing |
| Protein Glycosylation- Implication | It is the pattern and the chemical nature of the N-linked saccharides which exhibits a drastic change at the transition from moderate to extreme halophily. This different pattern of glycosylation (sulfated glucuronic acids and a repeating unit saccharide) introduces at least 120 additional negative charges into the glycoprotein. The protruding highly negatively charged loops are required for stabilization in high salt concentrations. Thus, the sulfated repeating unit saccharide is required for s |
| Glycan Information |
| Glycan Annotation | Linkages: βGalNAc- Asn, Glc-Asn, Gal-Thr.
Total carbohydrate content is approximately 10 to 12% of 200kDa S layer glycoprotein.
N glycosylated (at position N2) with GlcNAc-linked repeating (10-15 repeats) sulfated pentasaccharide and with sulphated oligosaccharides: (GlcA(OSO3-)-[β1→4GlcA(OSO3-)]2-β1→4Glc-Asn) at ten other poistions.
Abut 20 neutral di/ tri- saccharides α-D-Glc-(1→2)-Gal-(1→ or (uronic acid, glucose)-galactose are O-glycosidically attached to clustere |
| Technique(s) used for Glycan Identification | GC-MS (gas chromatography-mass spectrometry) analysis of peracetylated alditols. |
| Protein Glycosylation linked (PGL) gene(s) |
| OST Gene Name | aglB/stt3 |
| OST NCBI Gene ID | 5953829 |
| OST GenBank Gene Sequence | 5953829 |
| OST Protein Name | AglB/STT3 subunit |
| OST UniProtKB/ SwissProt ID | B0R4T2 |
| OST NCBI RefSeq | YP_001689095.1 |
| OST EMBL-CDS | CAP13747.1 |
| OST UniProtKB Sequence | >tr|B0R4T2|B0R4T2_HALS3 Oligosaccharyl transferase OS=Halobacterium salinarum (strain ATCC 29341 / DSM 671 / R1) GN=stt3 PE=4 SV=1
MSETTDASGLSAGLPSRALGALKDWYHVPVLGAVLAFMFWVRVQSWDNFLQNGEVYLSGN
DAYYHLRQVTYTVHHFPETMPFDPWTNFPHGTLAGQFGTLFDQILAAVALVVGMGSPSEH
TIAMTLLLAPPVFGALLVVPTYVMGRRIAGRLGGLFGAVILGLLPGYFLQRTLAGAADHN
GAEPLFMTLAVVGLMVALSVAEREKPVFEQFRAWDATGLRDVVGWSVLAGVATAVYMWVW
PPGVLLVGIFGAFFLVKLVGDYVTGTSPDHVAVVGAVSMTTTALLMFAPLGESGFGVSGF
SFLQPVFALGVAVGCVVLAWLARVFDERSLSPAAYVATVAGTLVVVVGALALVAPGFWSS
LQSNLLNYIGLSSTADTRTIGEAQPFLFRTGRYGLGMFGVVFLEYGTAFFSALVGAGAIL
LKPHLTSGSVRRIVGVAGATAVLAIVAVSPAVPAAIGGVVGLGGQLTALLIAAVVLAAIA
ATGQYDADKLLLVVWGAFVTSAAFTQVRFNYYLVVPVVVLNAYVLRAVLAAVDLDRPVAA
IEGVSWYQVGTVVIVALIVLVPVAAPAVAEATNNDSASGPVSPIKLSNNQNQPVPLQSAI
TAGQGQGPGGVVAWDDAMDWYQSHTPKQGTYGGADNADALDYNGTYSQTEDFDYPAGSFG
VLSWWDYGHWMTVQGHAIPHANPFQQGATSAANFLLADDEQQSESVLADIEEDDAKNRYV
AVDWKMVMPPLPYLSSGQSKFSAPVVFYDGPRDLSQGDFYQQAYNADWEENRISNARFFR
QSPLLKKQAYYESMMVRLYRYHGSAKQAEPVVADWRRSVNTGQGTAAAFDAATNNTKQFD
SMADAKAYVRNDSTA |
| OST EC Number (BRENDA) | 2.4.1.119 |
| OST Genome Context | |
| Characterized Accessory Gene(s) | |
| PGL Additional Links | CAZy |
| Literatures |
| Reference(s) | 1) Mengele, R. and Sumper, M. (1992) Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J Biol Chem, 267, 8182-8185. [PubMed: 1569073]
2) Lechner, J. and Sumper, M. (1987) The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J Biol Chem, 262, 9724-9729. [PubMed: 3036870]
3) Paul, G., Lottspeich, F. and Wieland, F. (1986) Asparaginyl-N-acetylgalactosamin |
| Additional Comments | First prokaryotic glycoprotein that was charcterized experimentally for the site of glycosylation as well as glycan attached. The features unique to N glycosylation in H salinarium are: the majority of glycans are linked via glucose instead of GlcNAc or GalNAc to the Asn in protein. Presence of sulfated oligosaccharides that bind to a C60-dolichol monophosphate carrier lipid. Protein associated glycans differ mainly in terminal sugars. S layer proteins of H salinarium and H. volcanii are the exa |
| Year of Identification | 1974 |
| Year of Validation | 1987 |


