|
|
Content Table:--
Description of MHCBN terms:
| Name of field |
Description |
| Anchor Position |
means the position of amino acids in peptide sequence crucial for binding of peptide with MHC allele.The numerical position
of anchor amino acid is determined by counting amino acid number from right to left with consideration that right most amino acid has "1" position |
| Peptide Dataset |
set of all peptide sequences interacting with specfic MHC allele in specified condition |
| Antigenic Blast |
Alingment tool developed for similarty search
of quary sequence against experimentally deteremined antigenic sequences
present in MHCBN |
| Binding affinity |
measure of binding affinity of peptide
with specific mhc allele.It is mostly determined by IC 50 Value |
| Database Reference |
Provide full information about the proteins of swissprot database
having sequence similiar to peptide sequence |
| Human Leukocyte
Antigen (HLA) |
Relating to histocompatibility antigens or the
major histocompatibility complex in human-often used with a letter to designate
a genetic locus and a number to designate an allele at the locus or the
antigen of the major histocompatibility complex corresponding to the locus
and the allele. |
| Host organism |
Host in which specfic mhc allele is occuring.eg
Human, mouse, monkey,chimpanzee etc |
| major histocompatibility
complex (MHC) |
A cluster of genes encoding polymorphic cell-surface
molecules (MHC class I and class II) that are involved in interactions
with T cells. These molecules also play a major role in transplantation
rejection. Several other non-polymorphic proteins are encoded in this region. |
| MHC Allele |
MHC alleles are intraspecies variance at particular
mhc gene locus.or in simple terms mhc alleles are diffrent forms of mhc
gene which diffre at particular gene locus |
| MHC Classes |
Two major mhc classes (1) MHC class-1 (2) MHC
class-2 HLA-A,B,C,G are major mhc class-1 alleles of human responsible
for endogenous antigen( self altered cells) processing and and
presentaion and HLA-DP,DR,DQ are major MHC class-2 alleles Responsible for
exogenous antigen processing and presentaion.In Mouse H-2Kd,H-2Db,H-2Dd
are major MHC class-1 alleles and I-Ak,I-Ab,I-Ek are major MHC class-2
alleles |
| Method |
means experimental method used for finding binding affinity and T cell activity of peptide. |
| MHC Blast |
Alingment tool developed for similarty search
of quary sequence against mhc allele sequences present in MHCBN |
| Source Protein |
Antigenic protein are those from which small
peptide having immunological properties obtained.This field also
provide information about start and end position of immunologcal
peptide. |
| Source of Information |
This field provide information about the
source from which the all information about particular entry of
database has been obtained.The information source can be public
database or published literature |
| T-cell Activity |
Semi-Quantitive measure of immunogenic potency of peptide. |
 Top of page Top of page
Retrieve help on General Query in MHCBN::
Stepwise Help:
Submit Query sequence
| Search
by entry number: Enter entry number [eg.15002] of peptide/record
in text box to obtain all the information about that entry in MHCBN. The
entry number must be of five digits.
|
|
Search by selecting MHC allele: Select one or more MHC allele from option box.The
default selected value for MHC allele is "All alleles". One can modify
query conditions by selecting the other search condition from
appropriate search option box. otherwise the default value for each of
option box used for search.Allele specific peptide with specific
binding affinity and T-cell activity can be obtain
by selecting the value of binding and T-cell activity from respective
option box. The user can obtain mhc allele specific non- binder peptides by
selecting the non-binder from binding affinity option box.Through general search
user can obtain peptides having no T cell activity even they are
mhc-binder by selecting "All" from binding affinity and "No" from T-cell activity.
|
|
Help for result field: user can restrict fields to be displayed in the result. All checked result
fields are displayed in result .if user did not select any check box then only the sequence corresponding
to selected condition will be displayed. Most of the fields in the
result are linked so that user can obtain realted information by just clicking
on any field value.All the publication references displayed in result
are hyperlinked to "pubmed".The reference database field also linked to
"swissprot And PIR" databanks to get relevent information about antigenic or
peptide sequences.The MHC allele sequence is linked to IMGT/HLA or ENTREZ.The
Antigenic structure field and MHC structure are linked to PDB.The result screen will show "No record
found under specific condition" when no relevant data found in database.
|
 Top of page Top of page |
Retrieve help for Peptide Search in MHCBN :: |
| This search option is
very useful for subunit vaccine desiging .This tool provide facility to overlapping
and msimatch peptide searches from database.The search also help to
elucidate the MHC interacting properties of query sequence.. A peptide
sequence binding with large number of interaspecific mhc alleles can be
a potential candidate for vaccine designing.A query peptide having high
binding affinity for many mhc alleles may be promocious T-cell epitope.This mismatch option of this tools allow search of such
peptides having few amino acid modified.
|
Stepwise help: |
| Paste or enter query
peptide sequence as single amino acid code in the provided text box.All the non
standard letters other than single AA code will be ignored.The " X "
character in the sequence will match with any amino acid.The sequence
of maximum 250 amino acids can be submitted. |
Example for Mismatch mapping:-
sample sequence: ILKEXXXPVHGV
This sequence will match with all the sequences from database which are similiar to query sequence except the xxx in sequence.
This sequence will match with
ILKEWQRPVHGV
ILKERQWPVHGV
ILKEKKKPVHGV
SADILKEPPPPVHGV
This way it match with many other sequence.
This sequence will not match with sequences which are differing from query sequence other than places having "X".
e.g.
QLKEWQRPVHGV
ILKEWQRPVHHV
ILYEWQRPVHHV
|
| Restricted Search: The
user can restrict search by selecting the value of mhc class,host organism,
binding affinity and T-cell activity from respective option box.The mhc
allele for which query peptide is non-binder can be obtained by selecting
non-binder option from Binding affinity.If user did not select any
option box then default value for each option box is considered for
search. |
| Help for result
fields:The fields in the result can be restricted by selecting
checkbox.The value of checked boxes will be displayed in result.The
defaut output only have Entry no.and MHC Allele corresponding to query
peptide according to selected
condition will be displayed.Every Field displayed in result is further
linked for obtaining more information. |
 Top of page Top of page |
|
Retrieve help on Dataset creation ::
|
This search option allow user to create datset for specific mhc allele
for specific condition. These datasets are very useful for designing of
prediction tools. |
Firstly Select MHC allele
from provided option box for which user want to create dataset.Restrict the
search by selecting the value of binding affinity and T-cell activity.The
unique dataset can be created for specfic mhc allele by selecting appropriate
checkbox.The result will show total number and list of all peptides.
Terminology Description
| Name of Major field | Name of sub field | Description |
| Peptide binding affinity |
ALL[Binders/Non Binders] |
All the binders and Non Binders |
| ,, |
ALL Binders |
All peptides with High, moderate or low binding affinity |
| ,, |
High |
Peptides with high binding affinity |
| ,, | Moderate | Peptides with moderate binding affinity |
| ,, | Low | Peptides with low binding affinity |
| ,, | Non Binders | Peptides with no binding affinity when tested in particular experimental condition. |
| Peptide T cell Activity | ALL | Peptides with known and unknown T cell Activity |
| ,, | ALL T cell epitopes | Peptides with known T cell Activity |
| ,, | High | Peptides with high T cell Activity |
| ,, | Moderate | Peptides with moderate T cell Activity |
| ,, | Low | Peptides with little T cell Activity |
| ,, | Non T cell epitopes | Peptides with no T cell Activity when tested in particular experiment |
|
Steps for creation of unique,datset of peptides with both high binding affinity and high T cell Activity for HLA-A*0201.
1.select "HLA-A*0201" from the list of MHC alleles provided in form.
2.select "HIGH" from binding affinity option box.
3.select "HIGH" from T-cell activity option box.
4.check "Unique Peptide Set for Specfic MHC Allele"
5.submit you query by clicking on "submit" button
Result page will show a dataset

 Top of page Top of page
|
|
Retrieve help on Peptide Mapping in Query sequence ::
|
This search option designed for mapping of MHC-binders,MHC non-binder and
T-cell epitope in query sequence.This tool allow user to map regions in query sequence which are suitable for vaccine designing. |
Stepwise help: |
Paste or enter your antigenic
or protein sequence as single amino acid code in provided text area.All
the non standard codes will be ignored from query sequence.User can restrict
search by selecting the value of host organism and type of peptide for
mapping. Otherwise the defaut value of each option box is considered for mapping.
Default value for host organism="All organisms".
Default value for type of peptide ="All". |
Help for result: The graphical (frames)
output is displayed in rows. The first row (black & bold)is amino acid
sequence(single amoino acid code) as submitted by the user. The remaining rows are
showing the location of mhc binders or non-binders or T cell epitopes
in the submitted sequence which are found in MHCBN.The different rows
or frames are taken in account for showing overlapping regions.The
maximum 20 frames are displayed for showing overlapping sequence.
Example:
1.Pasted query sequence:
"ilkepvhgvaaaaatruitutruittaaaaailkepvyioyiyyiyoyiy"
2.selected "Human" from mhc host organism.
3.keep default value for "MHC Binder" for type of petide.
4 submit the query.
This Type of Result are displayed.
|
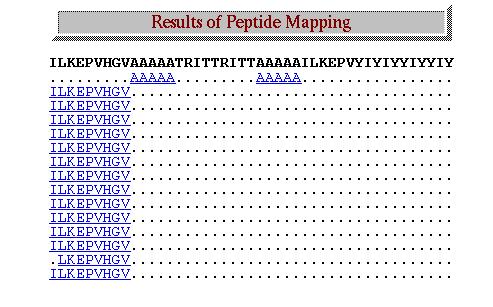
| Analysis
of result: Here in result the matching MHC binding regions are shown in
blue color. The dots (.) shows that non matching
sequence was found corresponding to that region of query sequence.The
further information about the MHC Binders can be obtained by just
clicking on it. |
 Top of page Top of page
Retreive help on
similarity search against Antigenic sequence :: |
| This option allows user to search
similar sequence in database of antigen proteins in MHCBN,
corresponding to their query search. Here, antigen proteins are those
proteins which consists of one or more peptides in MHCBN database. This
search is performed by BLAST program at GWBLAST server at IMTECH,
New Delhi.so more help can be obtained from stepwise help
|
Stepwise help: |
Pasting your sequence or Uploading a sequence file:
The Protein sequence can be pasted into the text area or a file containing
amino acids sequence can be uploaded using this option
|
Format Type:
In case the Format of the sequence is any of the standard ones (EMBL, FASTA, GENBANK, etc.)
then `Format Type' should be selected to `Standard Format (Readable by READSEQ)'. |
Choice for blast:
blastp - for protein - protein comparisons
|
Expectation value.
The lower the E value, the more significant the score. |
Select Region:
Users can select a particular region from the input sequence data for
similarity search. This option saves the user's job of continuously
trimming and editing their sequence in case where they want to restrict
their search to a particular region. |
Weight matrix:
BLAST uses different kinds of Substitution matrices for similarity
searches. It is well known that certain amino acids can substitute
easily for one another in related proteins, presumably because of their
similar physicochemical properties. These can be considered in
calculating alignment scores in a flexible manner through the use of a
substitution matrix, in which the score for any pair of amino acids can
be easily looked up.
Two most used matrices are PAM [Point accepted Mutation] and BLOSUM [BLOCKS substitution]
matrices.
PAM substitution matrices-
The first substitution matrices to gain widespread usage (Dayhoff et
al., 1978). It is a unit to quantify the amount of evolutionary change
in a protein sequence. 1 PAM unit is the amount of evolution which will
change 1% of amino acids in a
protein sequence. Although PAM250 was the only published PAM matrix,
the underlying mutation
data can be extrapolated to other PAM distances to produce a family of
matrices. When aligning sequences are highly divergent, best results
are obtained at higher PAM values,
such as PAM200 or PAM250. Matrices constructed from lower PAM values
can be used if the sequences have a greater degree of similarity.
BLOSUM subsitution matrices-
Constructed by Henikoff and Henikoff, 1992. The underlying data for BLOSUM are derived from
BLOCKS database (Henikoff and Henikoff, 1991), which contains local multiple alignments
("blocks") involving distantly related sequences. There is numbered series of BLOSUM matrices,
with the number referring to the maximum level of identity that sequences may have. e.g.
with BLOSUM62 matrix, sequences having at least 62% identity are merged into a single
sequence, so that the substitution frequencies are more heavily influenced by sequences that
are more divergent than this cutoff. |
For more help about antigenic BLAST 
|
 Top of page Top of page |
|
Retreive help on
similarty search against MHC allele sequence :: |
| This option allows user to search
similar sequences in database of MHC proteins in MHCBN, corresponding
to their query search. This search is
performed by BLAST program at GWBLAST.
|
|
The stepwise help for similarty search against MHC allele sequence
is just similiar to similarty search against antigenic sequence |
|
|

|



 Top of page
Top of page


