HLAncPred - Prediction of non-classical HLA Class-I binders
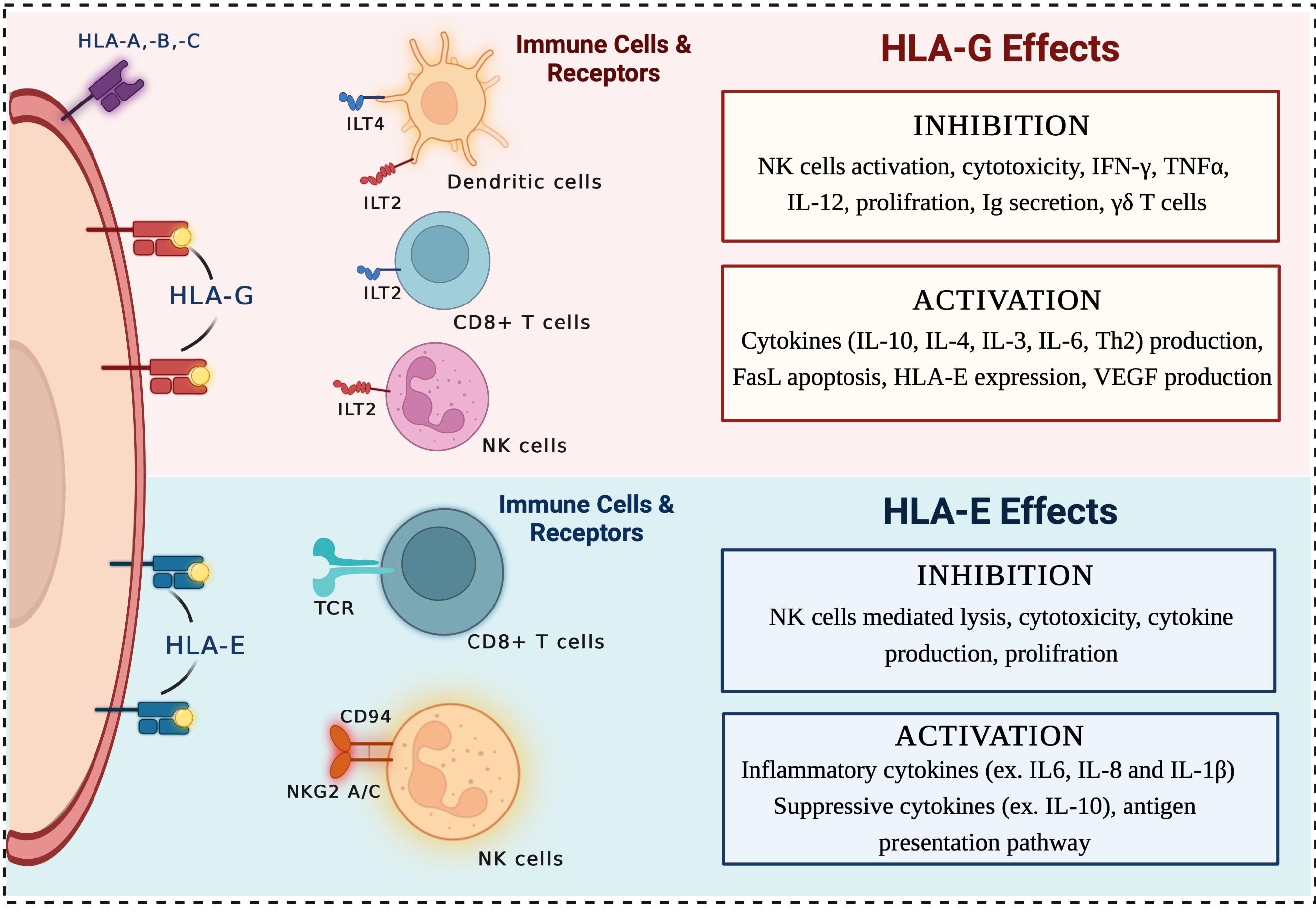
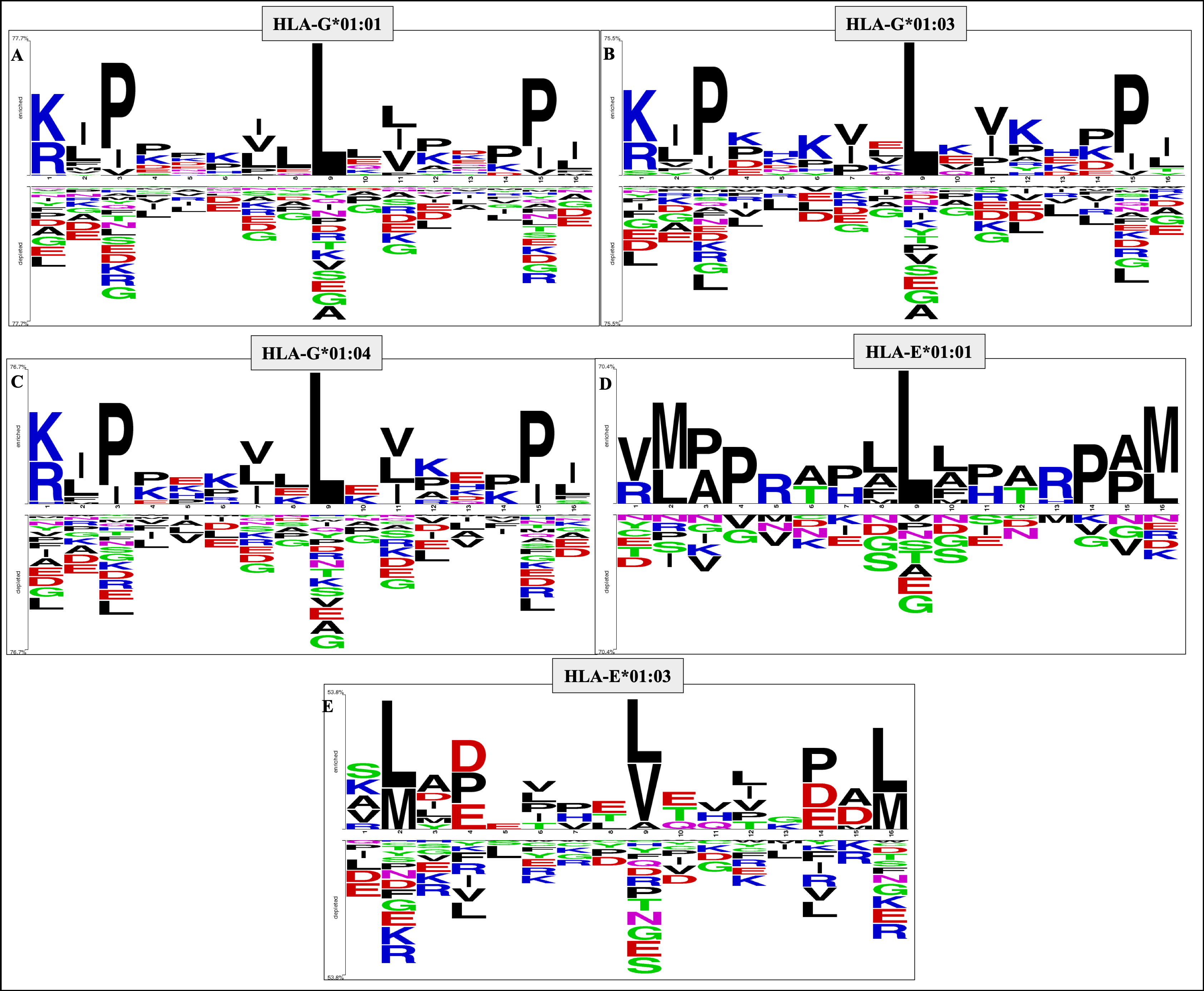
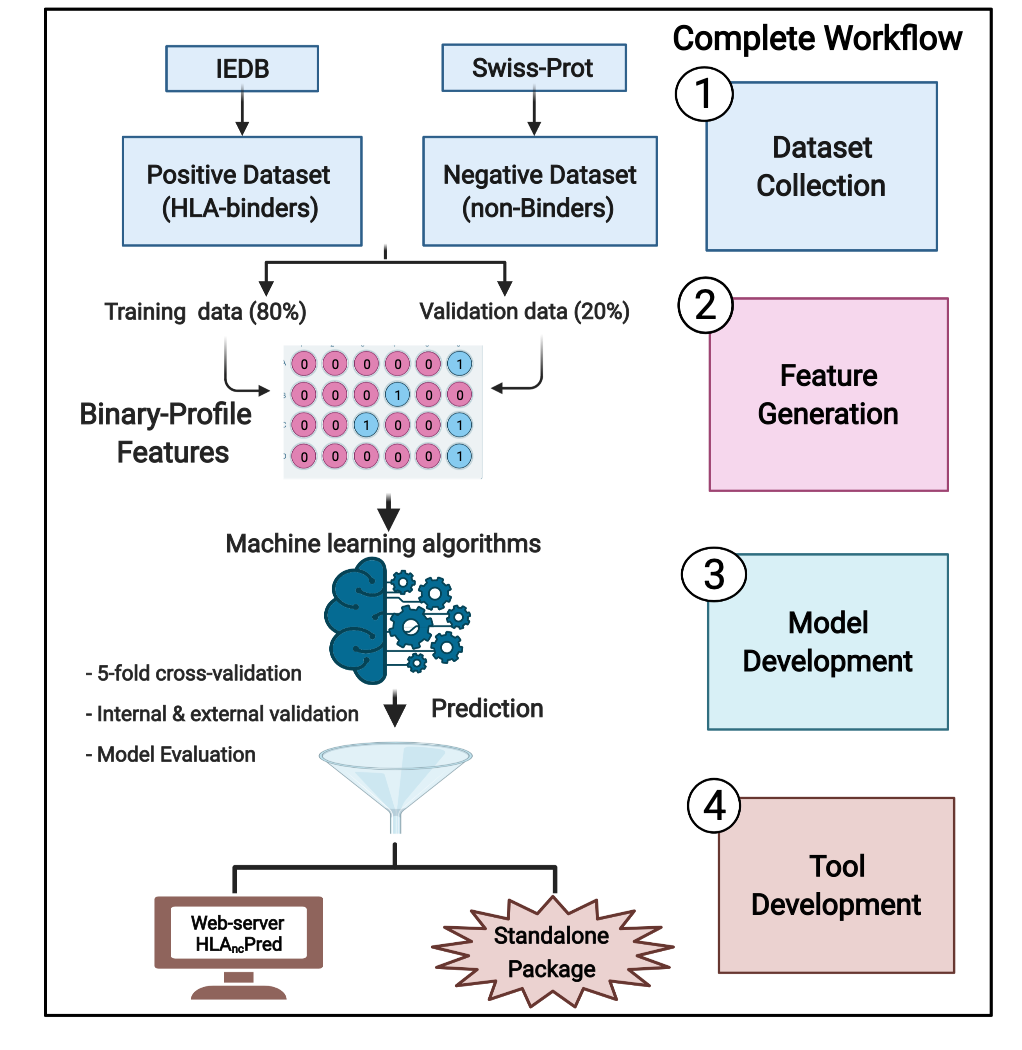
HLAncPred is a webserver for the prediction of promiscuous binders of non-classical HLA-G and HLA-E alleles (HLA-G*01:01, HLA-G*01:03, HLA-G*01:04, HLA-E*01:01 and HLA-E*01:03). In order to anticipate the scientific community we develop an in silico tool and package that allows the user to predict, and scan non-classical HLA binder/non-binder peptides.
Reference: Dhall A, Patiyal S, Raghava GPS (2022) HLAncPred: a method for predicting promiscuous non-classical HLA binding sites. Brief Bioinform. doi: 10.1093/bib/bbac192