The purpose of CHpredict server is to predict C-H...O and C-H...π interactions in a protein from amino acid sequence, particularly the interactions involving Cα atoms as hydrogen bond donors and oxygen atoms or aromatic ring as acceptors. It uses the amino acid sequence and secondary structure information for prediction. |
| Follow the following steps for prediction by CHpredict: |
| Step I: Paste the protein sequence in one-letter code in the textarea provided or upload the sequence file. Remember, the sequence should be in plain text format without any header line. |
| Step II: Choose the type of interaction to be predicted. It can be either Cα-H...O or Cα-H...π interaction or both. You will get an error if you leave this field blank. |
| Step III: You can enter your e-mail address at which you want to receive the results. This field is optional. you can also leave it as blank. |
| Step IV: Click on submit button to submit your prediction. |
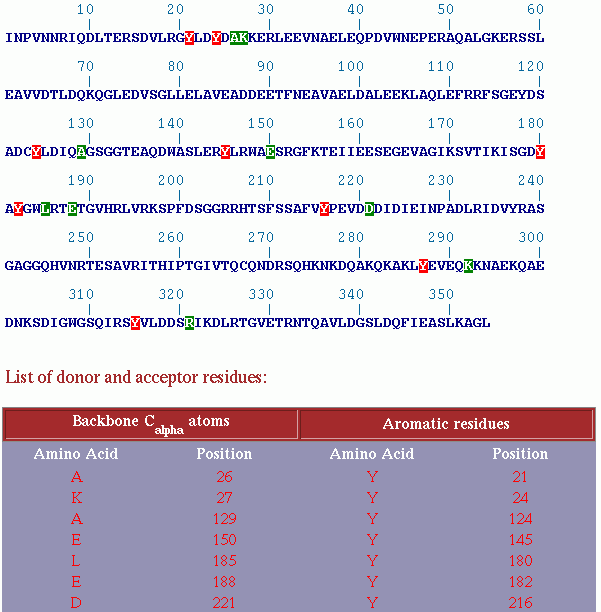
| The output consists of input sequence (sequence submitted by the user) where the donor (backbone Cα atoms) and acceptor residues involved in C&alpha-H...O interaction are marked in green and red color. Below the input sequence, the results are presented in tabular format consisting of donor and acceptor residues and their respective positions. |
 |
| The output consists of input sequence (sequence submitted by the user) where the donor (backbone Cα atoms) and acceptor (aromatic residues) involved in C-H...π interaction are marked in green and red color. Below the input sequence, the results are presented in tabular format consisting of donor and acceptor residues and their respective positions. |
 |
