Welcome to CancerPPD
|
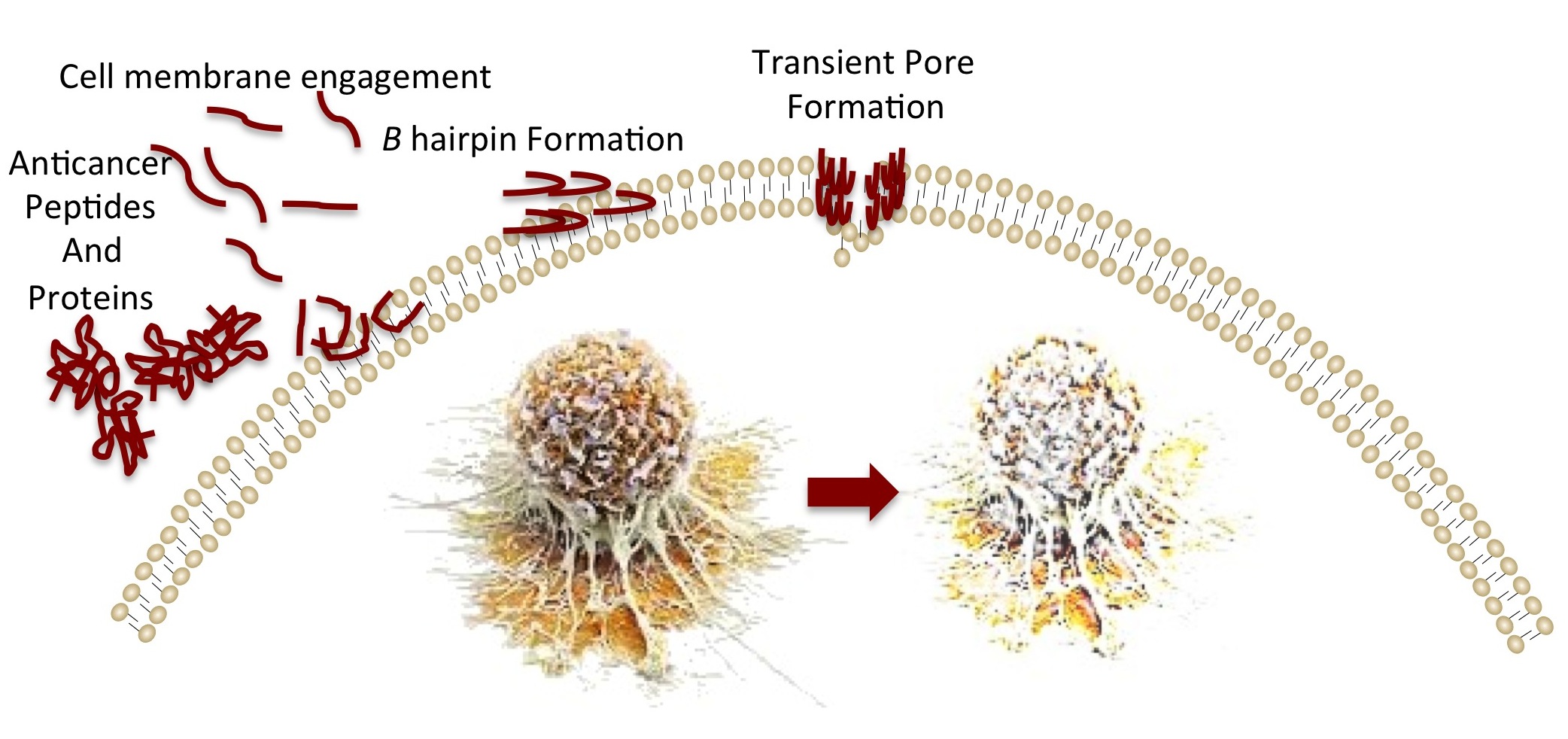
CancerPPD is a manually curated database of experimentally validated anticancer peptides. In this database, peptides have been collected from both published articles as well as from other repositories. In addition, tertiary structure of peptides have been predicted using PEPstr and secondary structure states are assigned using DSSP.Structure of modified peptides (containing Non-natural, D-amino acids, Modified-amino acid like Ornithine, Terminal modifications like Acetylation/Amidation) have also been predicted. In order to provide comprehensive information, peptides were searched and linked with important peptide and protein databases such as IEDB, PDB, Swiss-Prot and TrEMBL Major Features(1) Resource: It provides comprehensive information about hemolytic peptides that include their Sequence, Name, Origin, Type (Linear/Cyclic), Chirality, End modification, Chemical modification, Source,cytotoxic activity and Function. Data is collected from wide sources like literature and various other databases. Basic and Conditional Search facility enables the users to search a specific peptide/query in CancerPPD database.(2) Annotation: These peptides were searched in other databases to extract more information like their immunogenicity, source protein, functional annotation. (3) Web-tools Integrated: Various tools like BLAST, Mapping, Alignment have been integrated to facilitate users. (4) Data Retrieval: A number of data retrieval routines have been integrated to allow users to extract data from CancerPPD database that include search, browse, download routines. |