Algorithm
The prediction server for predicting the protein as allergenic and non-allergenic has been designed in a very user-friendly manner. This page explains the compatibility of webserver on different operating systems, details of all the algorithms and methods used to develop models for prediction. help page
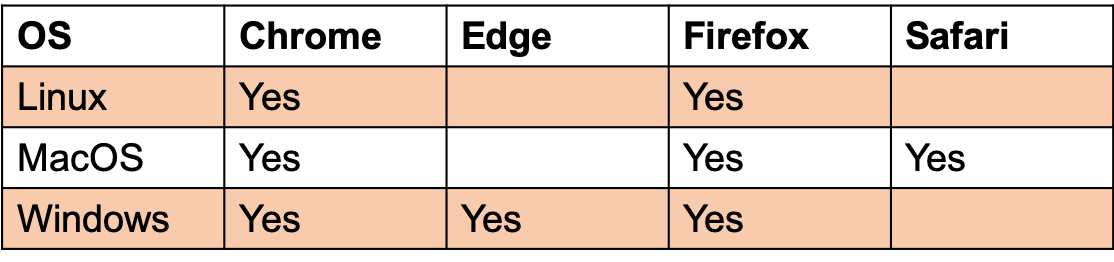
Browser compatibility:
Computation Time: Total computation time taken by AlgPred 2.0 for each protein in seconds (average)
Dataset collection and partitioning:
Firstly, we have collected the positive (allergen) and negative (non-allergen) data from five different sources: COMPARE, Allergen Online, AlgPred, AllerTop and Swiss-Prot. The number of allergens and non-allergens used in the study is 10,075. To create the dataset, we used CD-HIT at 40% sequence identity on both datasets and obtained clusters with similar sequences. The clusters were divided into training and validation dataset in 80:20 ratio. Training data was further divided into five sets to ensure non-redundancy.
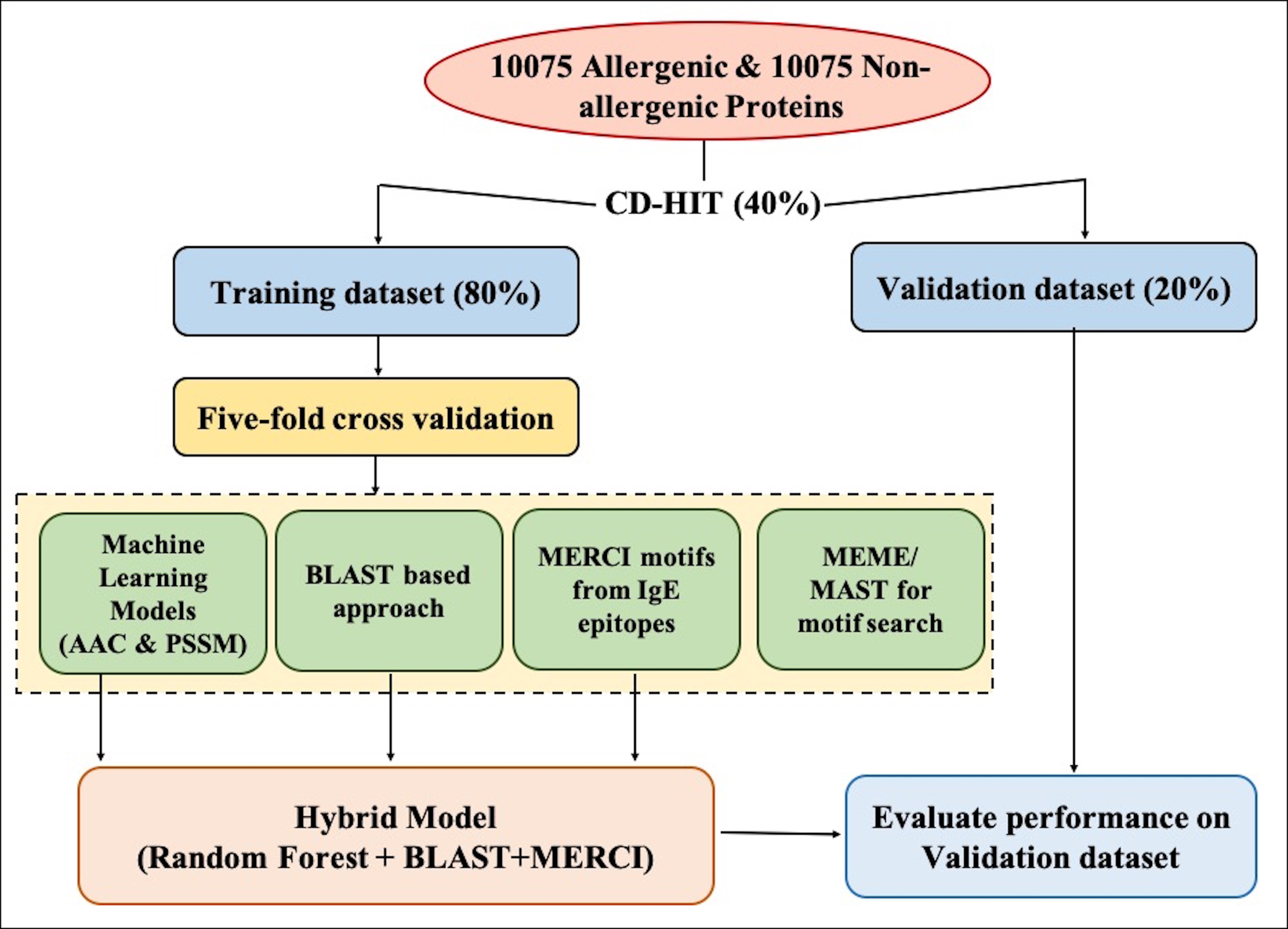
Machine Learning-based Model:
Different classifiers were used from Scikit-learn to develop the model. The software is freely available at https://scikit-learn.org/stable/
Random Forest (RF) based Model: The RF classifier is the best-performing model among others. It fits many decision trees and averages results to prevent over-fitting. Parameters such as n_estimators, min_samples_split, max_features can be tweaked.
Support Vector Machine (SVM) based Model: SVC classifier allows tweaking of kernel function like Linear, Polynomial, RBF, and Sigmoid.
K-Nearest Neighbor (kNN) based Model: Uses k-nearest neighbors to vote for each query point. Parameters such as n_neighbors, weights, and algorithm can be adjusted.
Multi-layer Perceptron (MLP) based Model: Builds models using neural networks. Parameters such as hidden_layer_sizes, activation, solver can be adjusted.
Protein Features Extraction
(a) Composition based features: Amino Acid Composition (AAC): 20-length vector representing fraction of each amino acid.
(b) Evolutionary information-based features: PSSM profiles: Generated using PSI-BLAST search against NCBI nr database. Vector length is 400 (20x20).
Five fold cross-validation: Dataset divided into five sets for training/testing; performance averaged over five iterations.
Performance Measure: Evaluated using threshold-dependent and threshold-independent measures.
Threshold-dependent Measures:
a) Sensitivity (Sens): Percentage of allergens correctly predicted.
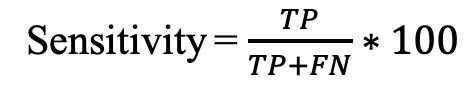
b) Specificity (Spec): Percentage of correctly predicted non-epitopes.
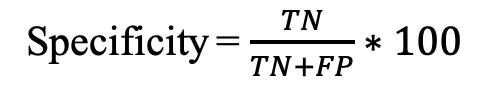
c) Accuracy (Acc): Percentage of total correct predictions.
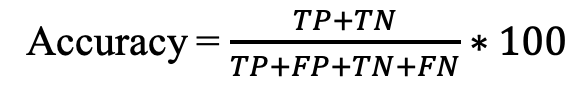
d) Matthews correlation coefficient (MCC): Correlation between observed and predicted values.
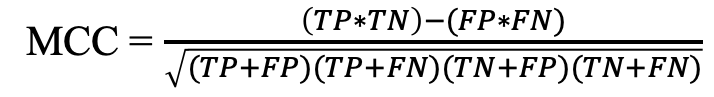
Where TP, TN, FP, and FN refer to true positives, true negatives, false positives, and false negatives.
Threshold-independent Measures: AUROC measures separability between classes.